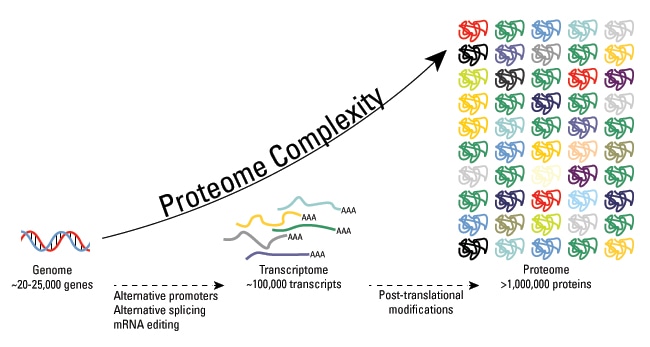
While the human proteome is encoded by approximately 20,000 genes, the functional diversity of the proteome is orders of magnitude larger because of added complexities such as genomic recombination, alternative transcript splicing, or post-translational modifications (PTMs). Post-translational modifications play a critical role in the regulation of protein structure, function, and cellular processes. As a leading company in the field of biological research and analysis, Creative Proteomics has been at the forefront of studying and understanding PTMs.
What are Post-translational Modifications?
Post-translational modification (PTM) refers to the covalent, usually enzymatic modification of proteins, and protein process during or after protein biosynthesis. Protein post-translational modification (PTM) increases the functional diversity of the proteome by the modifying proteins with functional groups, such as phosphate, acetate, amide groups, or methyl groups, and influences almost all the aspects of normal cell biology and pathogenesis. It plays a key role in many cellular processes such as cellular differentiation, protein degradation, signaling and regulatory processes, regulation of gene expression, and protein-protein interactions. The modifications genererally include phosphorylation, glycosylation, ubiquitination, nitrosylation, methylation, acetylation, lipidation and proteolysis and influence almost all aspects of normal cell biology and pathogenesis. Therefore, characterization of PTM, including the modification categories and modified sites, is critical in the study of cell biology and disease diagnostics and prevention.
Identification of Post-Translational Modifications is a tedious process. It can be affected by many factors. For example, most of the post-translational modifications are present in very low level. Therefore, enrichment steps are necessary before identification process. Additionally, stability of modification, and detection efficiency of mass spectrometry are also critical factors in PTMs identification process.
Select Service
Function of Post-Translational Modifications
Post-translational modifications play diverse and crucial roles in protein regulation and cellular processes. By modulating protein activity, protein-protein interactions, localization, and stability, PTMs finely tune cellular signaling networks and contribute to the complexity and adaptability of biological systems.
Modulating Protein Activity and Function
One of the primary roles of PTMs is to modulate protein activity and function. Phosphorylation, for example, acts as a molecular switch, turning proteins on or off by altering their enzymatic activity or subcellular localization. By adding or removing phosphate groups, kinases and phosphatases regulate signaling pathways and control cellular processes such as cell growth, proliferation, and apoptosis.
Glycosylation, on the other hand, affects protein structure and stability, influencing protein folding, solubility, and proteolytic resistance. It plays critical roles in protein-cell interactions, immune response, and cell adhesion processes. Ubiquitination, a PTM that tags proteins with ubiquitin molecules, regulates protein degradation by the proteasome, ensuring the removal of damaged or unwanted proteins and maintaining cellular homeostasis.
Modifying Protein-Protein Interactions
PTMs can also modulate protein-protein interactions, influencing cellular signaling cascades and molecular recognition events. N-acetylation and methylation, for instance, can impact protein-protein interactions by altering the charge distribution and conformation of protein surfaces. These modifications can facilitate or inhibit the binding of specific proteins or domains, regulating protein complex assembly, enzyme-substrate interactions, and signal transduction pathways.
Glycosylation, particularly N-linked glycosylation, plays a crucial role in mediating protein-protein interactions by serving as recognition motifs for carbohydrate-binding proteins. This enables glycosylated proteins to participate in cell-cell adhesion, immune response, and pathogen recognition.
Controlling Protein Localization and Trafficking
PTMs contribute to the precise localization and trafficking of proteins within cells. Phosphorylation, for instance, can trigger changes in protein conformation, leading to the exposure of nuclear localization or nuclear export signals. This regulates the shuttling of proteins between the cytoplasm and the nucleus, affecting their roles in transcriptional regulation, DNA repair, and cell cycle progression.
Glycosylation and lipid modifications, such as palmitoylation, myristoylation, and prenylation, facilitate the anchoring of proteins to specific cellular membranes, influencing their subcellular distribution and interactions with other membrane-associated proteins. These modifications play essential roles in signaling pathways, neuronal synapse formation, and vesicular trafficking.
Regulating Protein Stability and Degradation
PTMs are involved in regulating protein stability and degradation, ensuring proper turnover and quality control within cells. Ubiquitination, as mentioned earlier, tags proteins for proteasomal degradation. The addition of ubiquitin molecules to target proteins marks them for recognition by the proteasome, leading to their timely degradation and recycling of amino acids.
Other PTMs, such as acetylation and methylation, can impact protein stability by modulating interactions with chaperones or other regulatory proteins. These modifications can influence protein half-life and protect proteins from degradation or target them for proteolysis.
Post-translational modifications (PTMs) occur after protein synthesis and play crucial roles in expanding the functional diversity of proteins. These modifications introduce chemical changes to specific amino acid residues, altering protein structure, function, and behavior. Let's explore the mechanisms and processes involved in post-translational modification.
Enzymatic Machinery of PTMs
Post-translational modifications are catalyzed by specific enzymes that recognize target proteins and facilitate the addition or removal of chemical groups. For example, kinases and phosphatases are responsible for phosphorylation and dephosphorylation events, respectively. These enzymes transfer phosphate groups from ATP to specific amino acid residues or hydrolyze them, modulating protein activity and signaling pathways.
Glycosylation is mediated by glycosyltransferases, which transfer sugar molecules from activated sugar donors to target proteins. These enzymes recognize specific amino acid motifs and catalyze the attachment of sugars to appropriate residues, leading to the formation of glycosidic bonds.
Ubiquitination involves a cascade of enzymes, including E1 activating enzymes, E2 conjugating enzymes, and E3 ligases. The E1 enzyme activates ubiquitin molecules, which are then transferred to E2 enzymes. E3 ligases recognize specific target proteins and facilitate the transfer of ubiquitin from the E2 enzyme to the target protein, resulting in ubiquitin conjugation.
Recognition of Target Sites
PTMs occur at specific target sites within proteins, often determined by the presence of recognition motifs or consensus sequences. Enzymes involved in PTMs recognize these motifs and selectively modify the corresponding amino acid residues. For example, kinases recognize specific amino acid motifs surrounding serine, threonine, or tyrosine residues for phosphorylation. Similarly, glycosyltransferases recognize specific consensus sequences for glycosylation.
The recognition of target sites can be influenced by neighboring amino acids, protein conformation, and the presence of other PTMs. This complex interplay of factors determines the specificity and selectivity of PTMs, allowing for precise and regulated modification of proteins.
Regulatory Factors and Signaling Networks
Post-translational modifications are tightly regulated and integrated into complex signaling networks within cells. The addition or removal of PTMs is often triggered by specific cellular signals or environmental cues, leading to dynamic changes in protein function and cellular responses.
For example, phosphorylation events are often regulated by signaling cascades activated by extracellular ligands or intracellular stimuli. These signaling pathways involve multiple kinases and phosphatases, which are regulated by upstream signaling molecules and feedback mechanisms. The activation or inhibition of specific kinases or phosphatases can result in widespread changes in protein phosphorylation patterns, influencing cellular processes and gene expression.
Additionally, cross-talk between different PTMs can occur, where one modification can influence the occurrence or effects of another. For example, the phosphorylation state of a protein can affect its susceptibility to ubiquitination or alter the binding affinity of other PTM-regulating enzymes. These interactions further contribute to the complexity and regulation of post-translational modification networks.
What are the Most Common Post-translational Modifications?
Post-translational modifications (PTMs) are essential regulatory mechanisms that expand the functional repertoire of proteins. Phosphorylation, glycosylation, ubiquitination, N-acetylation, and methylation are among the most common PTMs, each playing unique roles in protein regulation and cellular processes.
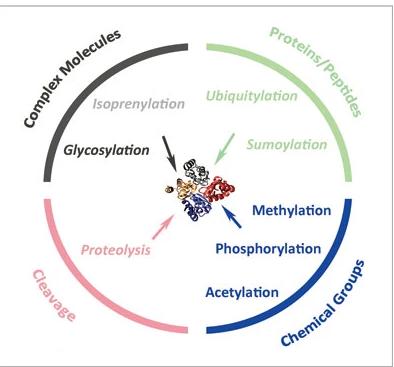
Types of post-translational modifications (PTMs)
Phosphorylation: Adding and Removing Phosphate Groups
Much of the activity in the cellular proteome is under the control of reversible protein phosphorylation. Phosphorylation-dependent signaling regulates differentiation of cells, triggers progression of the cell cycle, and controls metabolism, transcription, apoptosis, and cytoskeletal rearrangements. Signaling via reversible protein phosphorylation also plays a critical role in intracellular communication and immune response. Phosphorylation can function as a positive or negative switch, activating or inactivating enzymes. It can serve as a docking site to recruit other proteins into multiprotein complexes or serve as a recognition element to recruit other enzymes that add other post-translational modifications (PTMs) or additional phosphorylation sites. Phosphorylation can trigger a change in the three-dimensional structure of a protein or initiate translocation of the protein to another compartment of the cell.
Select Service
Learn more
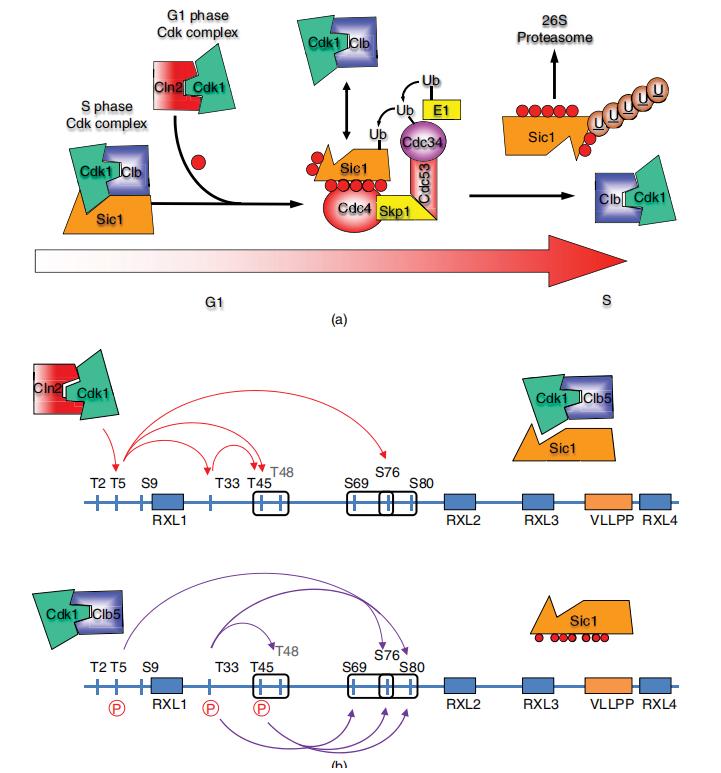
Cascades of multisite phosphorylation regulate biological function
Glycosylation: Adding Sugar Moieties
Glycosylation is the attachment of sugar molecules to proteins, resulting in glycoproteins. It is a highly diverse and complex post-translational modification that can occur in various forms. N-linked glycosylation involves the attachment of sugars to asparagine residues, while O-linked glycosylation occurs on serine, threonine, or hydroxylysine residues.
Glycosylation plays crucial roles in protein folding, stability, and cell-cell interactions. It contributes to protein quality control, protecting proteins from degradation. Moreover, glycosylation is involved in immune responses, cell adhesion, and the recognition of pathogens. The specific pattern and composition of glycans attached to proteins influence their functions and interactions with other molecules.
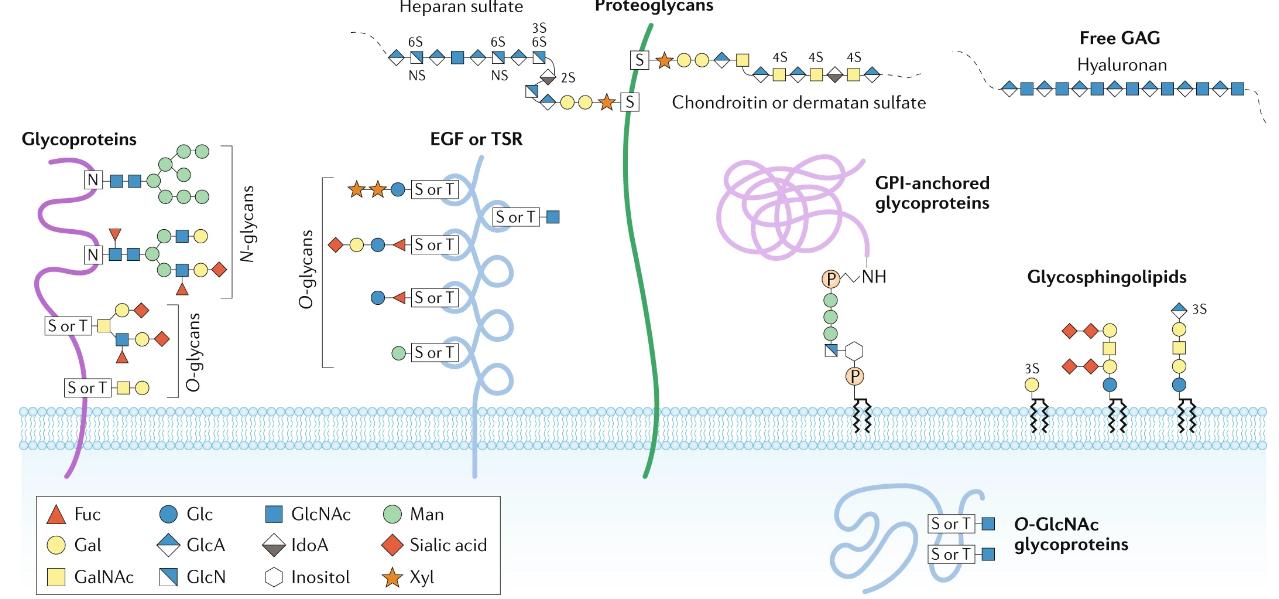
Major types of glycosylation in humans
Ubiquitination: Targeting Proteins for Degradation
Ubiquitination is a PTM that involves the covalent attachment of ubiquitin, a small protein, to target proteins. It acts as a signal for protein degradation by the proteasome, a cellular machinery responsible for protein turnover. Ubiquitination occurs through a series of enzymatic reactions involving E1 activating enzymes, E2 conjugating enzymes, and E3 ligases.
By tagging proteins with ubiquitin, ubiquitination regulates protein stability, turnover, and quality control. It targets damaged or unwanted proteins for degradation, ensuring proper cellular homeostasis. Additionally, ubiquitination plays roles in DNA repair, cell cycle regulation, and immune responses.
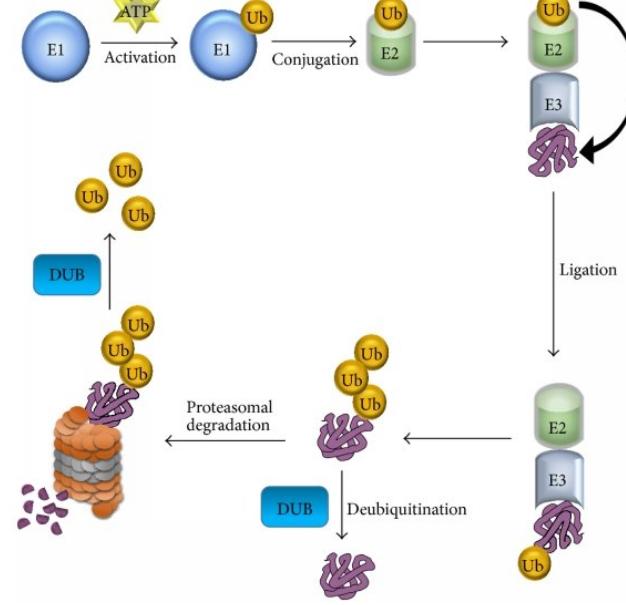
The ubiquitin proteasome system
Select Service
N-acetylation: Modification at the N-terminus
Protein acetylation are PTMs occurring predominantly on lysine residues. Acetylation can also occur at the N‐terminus and acts as multifunctional regulator including regulation of protein stability, via a specific degradation signal, termed the Ac/N‐degron. Originally considered epigenetic modifications that occurred mainly or exclusively on histones, it has become clear that acetylation also occur on a range of nonhistone proteins with modification rates that are subject to metabolic control. As such these PTMs can be considered key regulatory events in cell function.
Acetylation, as for other PTMs, can occur singly or in combination providing mechanisms for regulating protein activity through generating functional variants and regulatory sites. MS analysis of acetylation is directed not only to site identification and localization but also characterization of their combinations.
Select Service
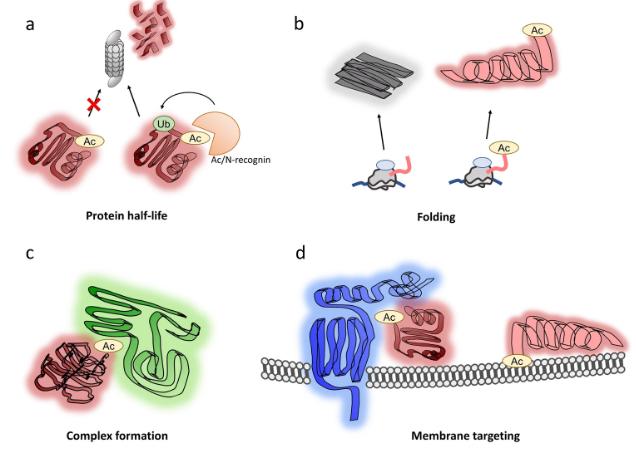
Functions of N-terminal acetylation.
Methylation: Adding Methyl Groups
Methylation is the addition of methyl groups to specific amino acid residues, most commonly lysine and arginine. It is catalyzed by methyltransferases, which transfer methyl groups from methyl-donating compounds to target amino acids. Methylation can occur on multiple sites within a protein, resulting in mono-, di-, or trimethylation.
Methylation plays diverse roles in protein regulation. It can influence chromatin structure, gene expression, and DNA repairprocesses. Methylation of histone proteins, for example, is involved in the epigenetic regulation of gene expression, where certain methyl marks are associated with gene activation or repression.
Methylation can also impact protein-protein interactions and protein stability. Methylation of arginine residues, for instance, can serve as docking sites for protein interaction domains, facilitating the assembly of protein complexes. Additionally, methylation of lysine residues in certain proteins can prevent their degradation by inhibiting recognition by ubiquitin ligases.
Understanding the diverse functions of these PTMs provides valuable insights into the complex regulatory networks within cells. Creative Proteomics, with its expertise in post-translational modification analysis, offers comprehensive solutions to study and analyze these modifications. By elucidating the role of PTMs, researchers can unravel the intricate mechanisms underlying protein regulation and cellular dynamics, ultimately advancing our understanding of biological systems.
Select Service
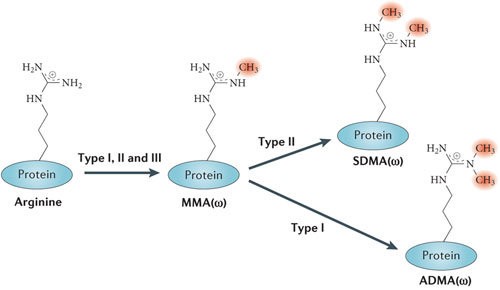
Types of methylation on arginine residues
Post-Translational Modification Detection Techniques
Accurate detection and analysis of PTMs are crucial for understanding their functional implications and their role in disease development. Creative Proteomics has been actively involved in developing and utilizing cutting-edge techniques for PTM detection. Here are some of the commonly used techniques in PTM detection:
Antibody-Based Techniques
Antibody-based techniques are widely employed for the detection of specific PTMs. Immunoblotting, also known as Western blotting, allows the identification and quantification of PTMs by utilizing antibodies that specifically recognize modified protein targets. This technique enables researchers to investigate the presence and abundance of specific PTMs across different samples or experimental conditions.
Immunoprecipitation (IP) is another valuable antibody-based technique used to isolate and enrich PTM-modified proteins from complex biological samples. By using antibodies specific to the PTM of interest, researchers can selectively capture the modified proteins and subsequently analyze them using various downstream techniques such as mass spectrometry.
Mass Spectrometry-Based Approaches
Mass spectrometers are powerful, analytical tools that have evolved rapidly over the past few decades to become the instrument of choice for protein and peptide characterization. Mass spectrometry is often used in parallel to other techniques such as western blot analysis or protein microarrays for detecting and quantifying PTMs. One of the main advantages of mass spectrometry is the ability to rapidly analyze many samples in a high‐throughput manner. Mass spectrometric analyses can be divided into three main strategies: “bottom‐up,” “middle‐down,” and “top‐down” proteomic approaches. Laboratories typically employ bottom‐up proteomic methodologies to characterize PTMs. Proteins of interest are purified and proteolytically digested with an enzyme such as trypsin, with resultant peptides being separated by reversed‐phase chromatography or another analytical method compatible with mass spectrometric analysis. One of several fragmentation methods and ion detection methodologies can then be employed. It is common to associate “data‐dependent” MS/MS analysis with bottom‐up approaches, where resulting peptide spectra are then pieced back together in silico to give an overview of the protein and its PTMs.
Learn more
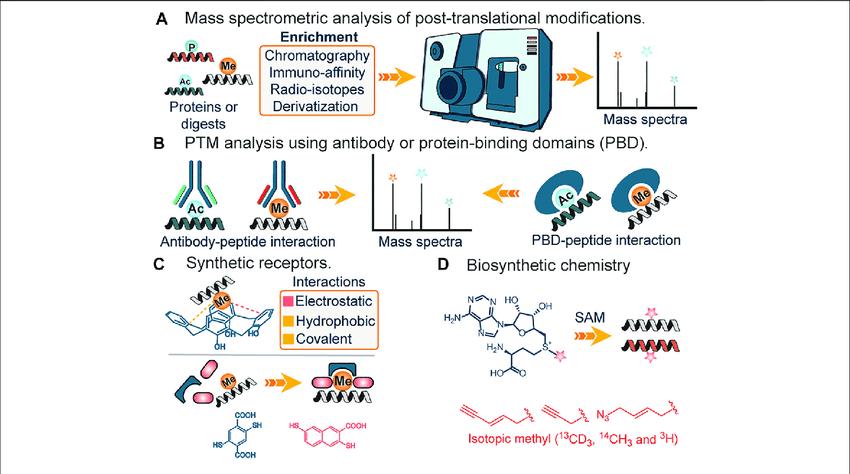
Analysis of PTMs using mass spectrometry.
Functional Assays
Functional assays are valuable tools for studying the impact of PTMs on protein activity and function. These sophisticated assessments offer unique perspectives into the intricacies of PTMs and how they shape the functions of proteins within cellular processes. Consider the high-powered and technologically advanced kinase assays, for instance, which enable researchers to determine the precise impact of phosphorylation on enzymatic activity. Such meticulous investigations are conducted by monitoring the transfer of phosphate groups to specific substrates, allowing for a theoretically limitless array of research possibilities.
Other functional assays include reporter gene assays, which evaluate the influence of PTMs on gene expression regulation, and enzyme activity assays, which assess the impact of modifications on catalytic activity. These assays allow researchers to link PTMs to specific functional outcomes and unravel the intricate regulatory networks controlled by PTMs.
Emerging Technologies
Advancements in technology have paved the way for the development of innovative approaches for PTM analysis. For instance, proximity-dependent labeling techniques, such as BioID and APEX, enable the identification of PTM-regulated protein-protein interactions in a spatially resolved manner within living cells. These techniques rely on the proximity of a modified protein to a promiscuous biotin ligase, resulting in the biotinylation of nearby interacting proteins.
Intriguingly, recent developments in the world of scientific research have given rise to cutting-edge screening methods, referred to as high-throughput screening approaches. Such advanced techniques include peptide microarrays and protein arrays, and hold within them the unparalleled potential to explore the complex intricacies of PTM-binding proteins and the recognition patterns of PTM-specific antibodies. With their emergence comes a newfound opportunity to comprehensively analyze PTMs, vastly expanding our understanding of the biological processes they mediate.
What Can We Do for Your PTMs Research
Post-translational modifications are essential regulatory mechanisms that govern protein function and cellular processes. Understanding the intricacies of PTMs requires comprehensive analysis techniques that can detect, quantify, and characterize these modifications. Creative Proteomics, with its expertise in post-translational modification analysis, employs a range of techniques such as antibody-based approaches, mass spectrometry, and functional assays to unravel the functional implications of PTMs.
Through the utilization of these techniques, Creative Proteomics aims to contribute to the advancement of knowledge in the field of post-translational modification analysis, ultimately leading to a deeper understanding of PTMs' roles in cellular signaling, disease mechanisms, and therapeutic interventions. As a leader in the industry, Creative Proteomics remains dedicated to pushing the boundaries of post-translational modification analysis and delivering innovative solutions to researchers and scientists.
Types of PTMs analysis service we provide:
- Acylation Analysis
- Acetyl-proteomics
- Alkylation Analysis
- Biotinylation Analysis
- Crotonylation Analysis
- Di-Sulfide Bond Localization
- Glutathione Sites Identification
- Glycosylation Analysis
- Glutamylation Analysis
- Hydroxylation Site Identification
- Histone PTMs Analysis
- Lipidation Analysis
- Methyl-proteomics
- Propionylation Analysis
- Phosphoproteomics
- Palmitoylation Analysis
- S-Nitrosylation Analysis
- S-myristoylation Analysis
- S-prenylation Analysis
- Succinylation Analysis
- SUMOylation Analysis
- Ubiquitinated-proteomics
Workflow of our PTMs analysis service:
- Digestion of proteins into small fragments
- Protein separation and analysis using LC/MS/MS
- Database search
- PTM mapping
- Full protein annotation
Technology platform:
- 2D Fluorescence Difference Gel Electrophoresis (DIGE)
- Liquid Chromatography (LC)
- High Performance Liquid Chromatography (HPLC)
- Matrix Assisted Laser Desorption Ionization Mass Spectrometry (MALDI-MS)
Advantages of our PTMs Analysis:
- Newest version of mass spectrometer: Q ExactiveTM MS/MS, with the highest resolution.
- Wide and full coverage range: at least 3 enzymes will be used to ensure wide and full coverage.
- Professional data analysis: constantly upgraded bioinformatics analysis systems.
Ordering Procedure:
References
- John R. Griffiths and Richard D. Unwin. Analysis of Protein Post-Translational Modifications by Mass Spectrometry. 2017
- Wang, YC., Peterson, S. & Loring, J. Protein post-translational modifications and regulation of pluripotency in human stem cells. Cell Res 24, 143–160 (2014).
- Reily, C., Stewart, T.J., Renfrow, M.B. et al. Glycosylation in health and disease. Nat Rev Nephrol 15, 346–366 (2019).
- Ree, R., Varland, S. & Arnesen, T. Spotlight on protein N-terminal acetylation. Exp Mol Med 50, 1–13 (2018).
- Yang Y, Bedford M T. Protein arginine methyltransferases and cancer. Nature Reviews Cancer, 2013, 13(1): 37.