Integrated Metabolomics and Microbiomics Analysis
Creative Proteomics specializes in integrated metabolomics and microbiomics analysis, delivering precise, high-throughput multi-omics solutions. We provide comprehensive services including untargeted and targeted metabolomics, 16S rRNA sequencing, shotgun metagenomics, SCFA profiling, lipidomics, and host-microbiota co-metabolome analysis. Our platform enables in-depth exploration of host-microbiota interactions, microbial functions, and metabolic pathways, helping clients accelerate research in health, agriculture, environment, and biotechnology with highly reproducible, customizable data outputs.
Submit Your Request Now
×- What We Provide
- Advantage
- Workflow
- Technology Platforms
- Sample Requirements
- FAQ
- Publication
Why is Integrated Metabolomics and Microbiomics Analysis Necessary?
The microbiome is the collection of genetic material from all microorganisms inhabiting a given environment. Microbiome analyses can identify differences in colony structure and abundance, and predict or annotate differences in colony function. The metabolome represents all small molecules present in the same environment, and it reflects colony-host interactions. Both the microbiome and metabolome play critical roles in regulating host health, disease development, and ecosystem dynamics. Integrated microbiome and metabolome analyses can help understand how microflora influence the metabolic state of the host through colony metabolism and co-metabolism with the host, and can be used to gain insights into microbial interactions and functions in a system.
Creative Proteomics is an industry leader in multi-omics analysis. In integrated metabolome and microbiome analysis, we analyze the microbiome and metabolome separately, and then use multi-omics analysis technology to correlate the microbiome data with the metabolome data. Integrated metabolome and microbiome analysis can investigate the mechanisms of disease development, microbial-plant and animal-plant interactions. etc.
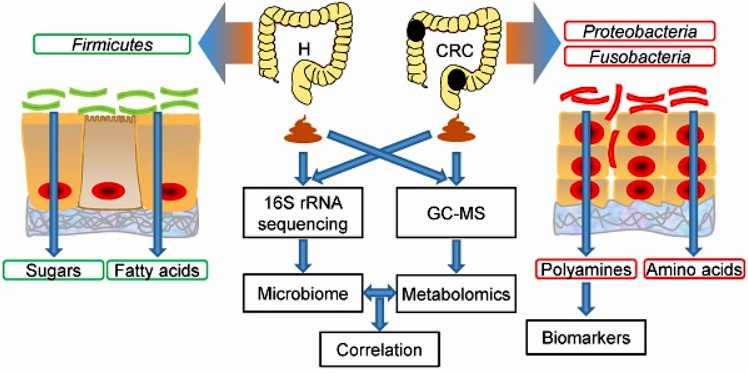 Integrated microbiome and metabolome analysis reveals a novel interplay between commensal bacteria and metabolites in colorectal cancer. (Yang Y, et al. 2019)
Integrated microbiome and metabolome analysis reveals a novel interplay between commensal bacteria and metabolites in colorectal cancer. (Yang Y, et al. 2019)
Comprehensive Integrated Metabolomics and Microbiomics Analysis Services Offered by Creative Proteomics
| Service Module | Description |
|---|---|
| Untargeted Metabolomics + 16S rRNA Sequencing | Simultaneous profiling of global metabolites and bacterial community composition. |
| Targeted Metabolomics + Shotgun Metagenomics | Quantitative analysis of selected metabolites along with high-resolution microbial gene function profiling. |
| Short-Chain Fatty Acids (SCFAs) Analysis + Microbiome Profiling | Specialized detection of SCFAs, key mediators of gut microbiota metabolism, integrated with microbiota structure data. |
| Lipidomics + Microbiomics | Integrated lipid profiling and microbiome analysis to explore lipid-microbiome interactions in metabolic diseases. |
| Host-Microbiota Co-Metabolome Analysis | Comprehensive evaluation of metabolites derived from host and microbiota metabolic interactions. |
| Multi-Omics Integration Consulting | Customized consulting services to design, execute, and interpret complex integrated multi-omics studies. |
Advantages of Integrated Metabolomics and Microbiomics Assay
- Full-Spectrum Biological Profiling: Simultaneously detect 1,000+ metabolites and 10,000+ microbial species per sample for comprehensive host-microbiota interaction insights.
- Ultra-High Sensitivity and Precision: Orbitrap LC-MS and NovaSeq™ achieve metabolite detection limits of 1 nM and microbial detection at 0.001% relative abundance.
- Robust Quantitative Integration: Integrated pipelines deliver metabolite–microbiome correlation with Pearson r > 0.9 and metabolomics CV < 15%, ensuring data reliability.
- Wide Sample Compatibility: Supports 10+ sample types (e.g., feces, plasma, tissue) with minimal input: 100 mg or 500 µL.
- Accelerated Turnaround: Delivers full reports within 6–8 weeks.
- Advanced Bioinformatics Visualization: Provides 30+ customized visual outputs, including metabolic pathways and microbe-metabolite network maps.
- High Data Reproducibility: Achieves >95% consistency for metabolites and >97% for microbial profiles across technical replicates.
- Flexible Customization: Over 90% of projects are fully customized, from target selection to analysis workflows.
Workflow for Integrated Metabolomics and Microbiomics Analysis Service
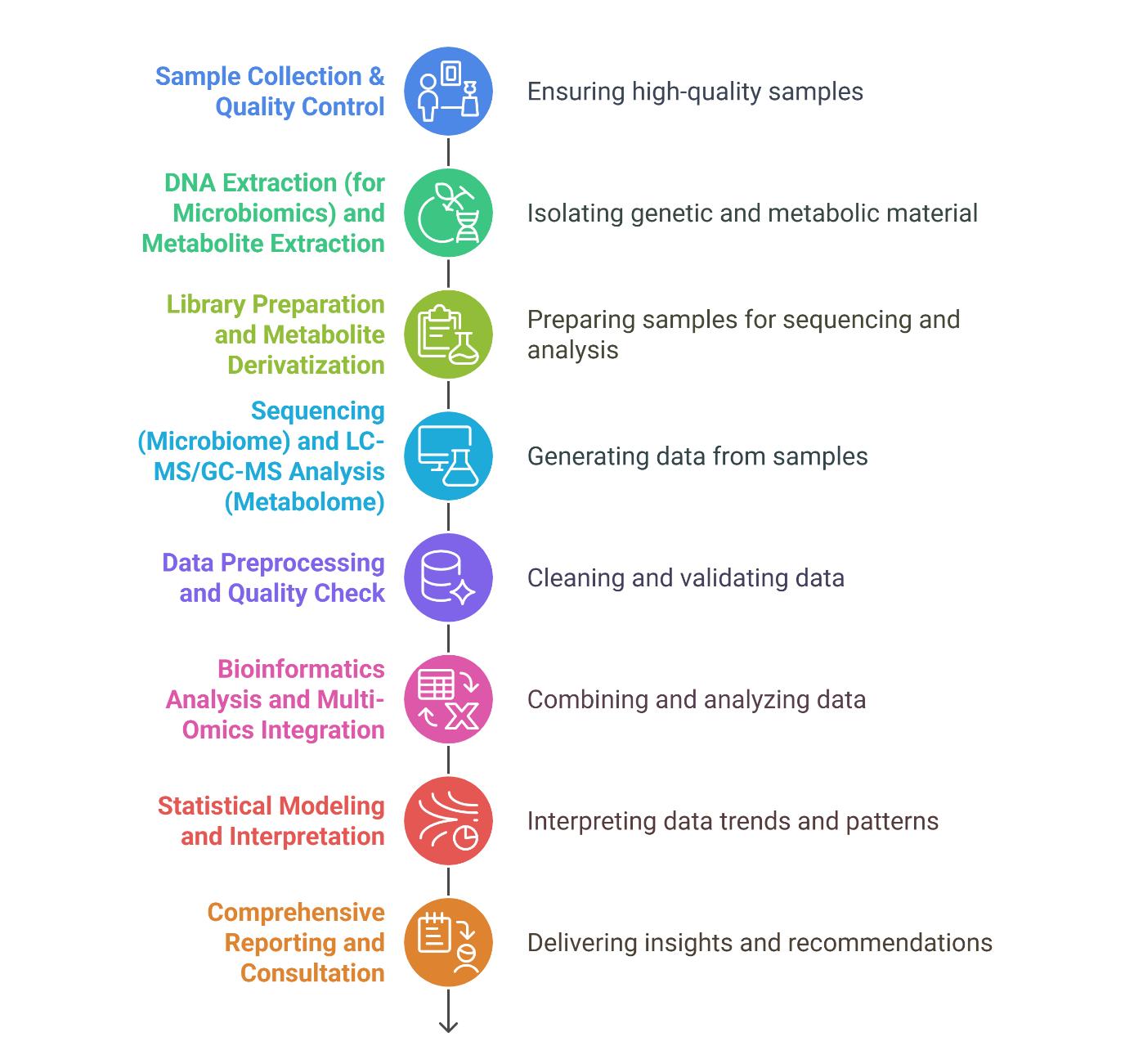
Technology Platform for Integrated Metabolomics and Microbiomics Analysis

Agilent 6495C Triple quadrupole (Figure from Agilent)

SCIEX Triple Quad™ 6500+ (Figure from Sciex)

Agilent 1260 Infinity II HPLC (Fig from Agilent)
Sample Requirements for Integrated Metabolomics and Microbiomics Analysis Service
| Sample Type | Required Volume/Amount | Notes |
|---|---|---|
| Plasma / Serum | ≥ 100 µL | Use EDTA or heparin tubes; avoid hemolysis |
| Urine | ≥ 500 µL | First-morning urine preferred; sterile container |
| Feces | ≥ 200 mg | Fresh or frozen; freeze immediately at −80°C |
| Tissue (e.g., liver, colon) | ≥ 100 mg | Snap-frozen in liquid nitrogen; no RNA later |
| Cell Pellet | ≥ 1 × 10⁶ cells | Wash with PBS, freeze immediately |
| Swab Samples (oral, vaginal, skin) | 1 swab per sample | Use sterile swabs; freeze at −80°C after collection |
| Intestinal Contents | ≥ 200 mg | Fresh or snap-frozen immediately |
| BALF (Bronchoalveolar Lavage Fluid) | ≥ 500 µL | Collect under sterile conditions; freeze immediately |
| Saliva | ≥ 500 µL | Rinse mouth with water before collection; no food 30 min prior |
Applications of Integrated Metabolomics and Microbiome Assay Service
Food and Nutrition Research
Reveals the impact of diet on host metabolism and gut microbial function to guide food innovation and nutritional optimization.
Agricultural Science
Analyzes soil, plant, and rhizosphere microbiome-metabolome interactions to enhance crop productivity and resilience.
Environmental Science
Investigates ecosystem health by profiling metabolite signatures and microbial communities in water, soil, and air samples.
Industrial Biotechnology
Optimizes microbial fermentation processes by linking metabolite profiles with microbial community dynamics.
Animal Health Research
Explores the relationship between host metabolism, microbiota, and overall animal health for feed additive development.
Cosmetic Research
Examines skin microbiota and metabolic activity to develop microbiome-friendly cosmetic formulations.
FAQ of Integrated Metabolomics and Microbiome Analysis Service
How should I handle samples for cross-omics studies to ensure data compatibility?
Use identical aliquots for metabolomics and microbiome sequencing. Avoid freeze-thaw cycles (>3 cycles degrade metabolite integrity) and store samples at -80°C immediately. For SCFA analysis, avoid lyophilization due to volatility.
What is the minimum biological replication required for statistically robust correlations?
- Animal studies: ≥10 replicates/group.
- Clinical studies: ≥30 replicates/group. Smaller cohorts risk false discoveries (e.g., Spearman correlation |r| >0.7 requires n=10 for 80% statistical power).
How do I choose between LC-MS and GC-MS for metabolomics in microbiome studies?
- LC-MS (Q Exactive HF-X): Ideal for polar/nonvolatile metabolites (e.g., lipids, bile acids).
- GC-MS (Agilent 8890): Superior for volatile compounds (e.g., SCFAs) with <5% CV reproducibility. Use untargeted LC-MS + targeted GC-MS for broad coverage.
Can archived samples (>2 years old) be used for integrated analysis?
Yes, but validate metabolite stability via QC modules (e.g., ISTD recovery rates ≥85%). Microbial DNA remains stable if stored at -80°C without thawing.
How are microbiome-metabolite correlations visualized and interpreted?
Tools include SparCC networks (filtering |r| >0.5, p<0.05) and heatmaps (hierarchical clustering). Prioritize interactions with Random Forest VIP scores >1.5 for biological relevance.
What bioinformatics methods integrate multi-omics datasets effectively?
MOFA+: For unsupervised latent factor analysis. Both require ≥90% feature concordance across datasets.
How to address batch effects in multi-omics projects spanning months?
Include interleaved QC samples (pooled biological replicates) in each batch. Use ComBat or SVA for cross-batch normalization.
Can fecal and blood samples from the same subject be analyzed together?
Yes, but interpret cautiously. Fecal metabolomes reflect gut microbiota activity, while blood metabolomes represent systemic absorption. Use multi-block PLS to model compartment-specific interactions.
How to validate putative microbiome-driven metabolic pathways?
Combine ex vivo fermentation assays (e.g., fecal cultures + isotope-labeled substrates) with qPCR (e.g., butyrate kinase gene buk) to confirm pathway activity.
Learn about other Q&A.
Integrated Metabolomics and Microbiome Analysis Service Case Study

Integrated fecal microbiome-metabolome signatures reflect stress and serotonin metabolism in irritable bowel syndrome
Journal: Gut Microbes
Published: 2022
- Abstract
- Results
- Conclusion
- Reference
The gastrointestinal disorder irritable bowel syndrome (IBS) is a classic example of microbiome-gut-brain axis involvement. The complexity of the underlying etiology of IBS, the involvement of psychosocial comorbidities, and its heterogeneity have proved to be a hurdle in the search for biomarkers and in the development of more effective therapeutic strategies. There are significant differences in IBS patients' gut microbiota characteristics compared to healthy controls (HC). However, there is still a lack of consensus on the exact nature of the changes in the gut microbiota composition in IBS. In addition to microbial composition, available data on microbial metabolic activity are limited and uncertain. Host-microbiome interactions may be largely driven by metabolic and microbial processes. Furthermore, knowledge of how changes in the gut microbiome and metabolome in IBS in particular reflect underlying pathophysiologic mechanisms or subgroups of IBS patients is very limited in this heterogeneous disease, and studies evaluating these pathways are needed to improve our understanding of host-microbiome interactions in IBS.
In the present study, the authors used whole-genome shotgun metagenomic sequencing (MGS) of gut microbiota composition combined with data on gut microbiota metabolic activity using proton nuclear magnetic resonance (1 H-NMR) spectroscopy to demonstrate that IBS microbiome-metabolome profiles are associated with altered serotonin metabolism and unfavorable stress responses associated with gastrointestinal symptoms, supporting the microbiota-gut-brain in the IBS pathogenesis link.
Based on H-NMR spectra, 50 unique metabolites have been identified in the fecal water of IBS patients and HC. Based on these metabolites, it was possible to differentiate between HC and IBS patients with an AUC of 79.5%. The metabolites responsible for this differentiation were increased in 10 metabolites in HC and 16 metabolites in IBS patients (Figure 1).
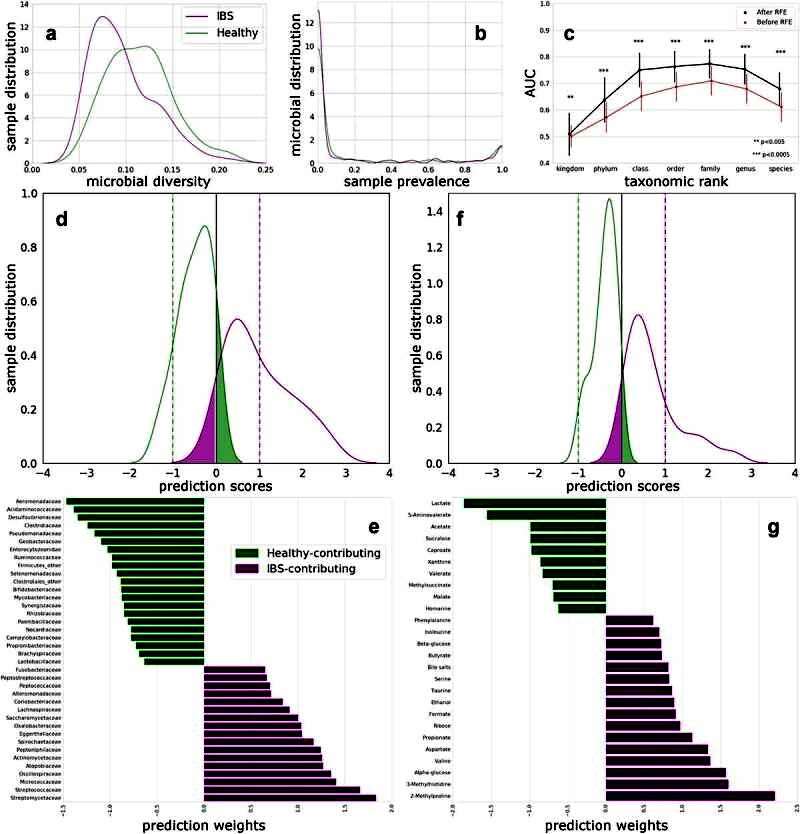 Figure 1 Distinguishing HC and IBS using microbiome at different taxonomic ranks, and using fecal water metabolites.
Figure 1 Distinguishing HC and IBS using microbiome at different taxonomic ranks, and using fecal water metabolites.
When combining fecal microbiota and fecal metabolites at the family level, it was possible to differentiate the two groups with an AUC of 83.6%. The model consists of microbial families and metabolites that distinguish HC and IBS patients. As shown in Figure 2c the combination of the two histologic levels provided the best differentiation between HC and IBS patients compared to using only the microbiota or fecal metabolites to differentiate the groups (Figure 2).
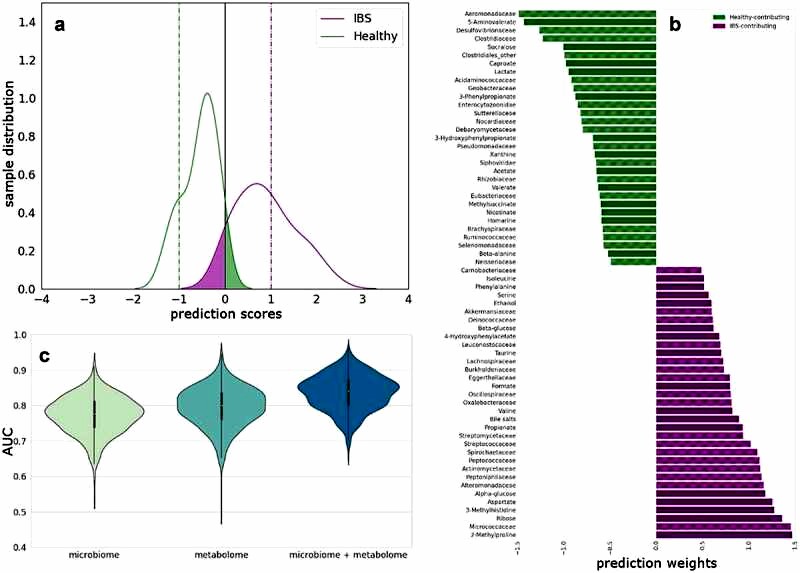 Figure 2 Multi-omics integration to predict differences between HC and IBS.
Figure 2 Multi-omics integration to predict differences between HC and IBS.
When considering IBS patients only, different patient clusters can be identified based on fecal microbiota and metabolites separately and in combination with both histologic data. These clusters are then investigated based on differences in the extensive clinical and biological metadata available in this cohort (Figure 3)
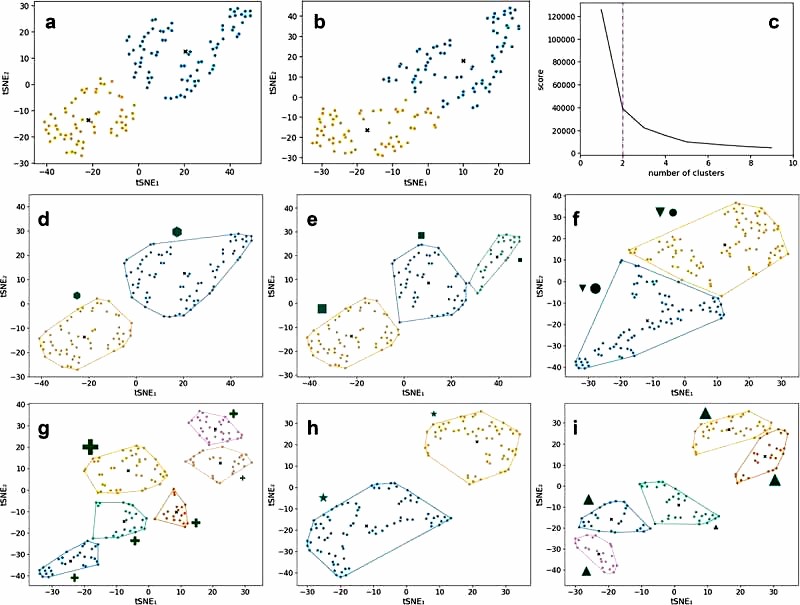 Figure 3 Unsupervised clustering using kernel t-SNE (kt-SNE)
Figure 3 Unsupervised clustering using kernel t-SNE (kt-SNE)
To determine the link between the gut microbiota and corresponding metabolites, an extensive metabolic response network was constructed involving gut microbial families and fecal metabolites significantly increased and decreased in patients with IBS. In this interaction diagram, microbial families with increased IBS (purple) and increased HC (green) are shown on the left. The pathways of fecal water metabolites can be traced on the right side of the graph by concentrating on enzymes; again, purple indicates an increase in IBS and green indicates an increase in HC (Figure 4). The pathways of glycolytic and proteolytic metabolic activity can be inferred from this figure.
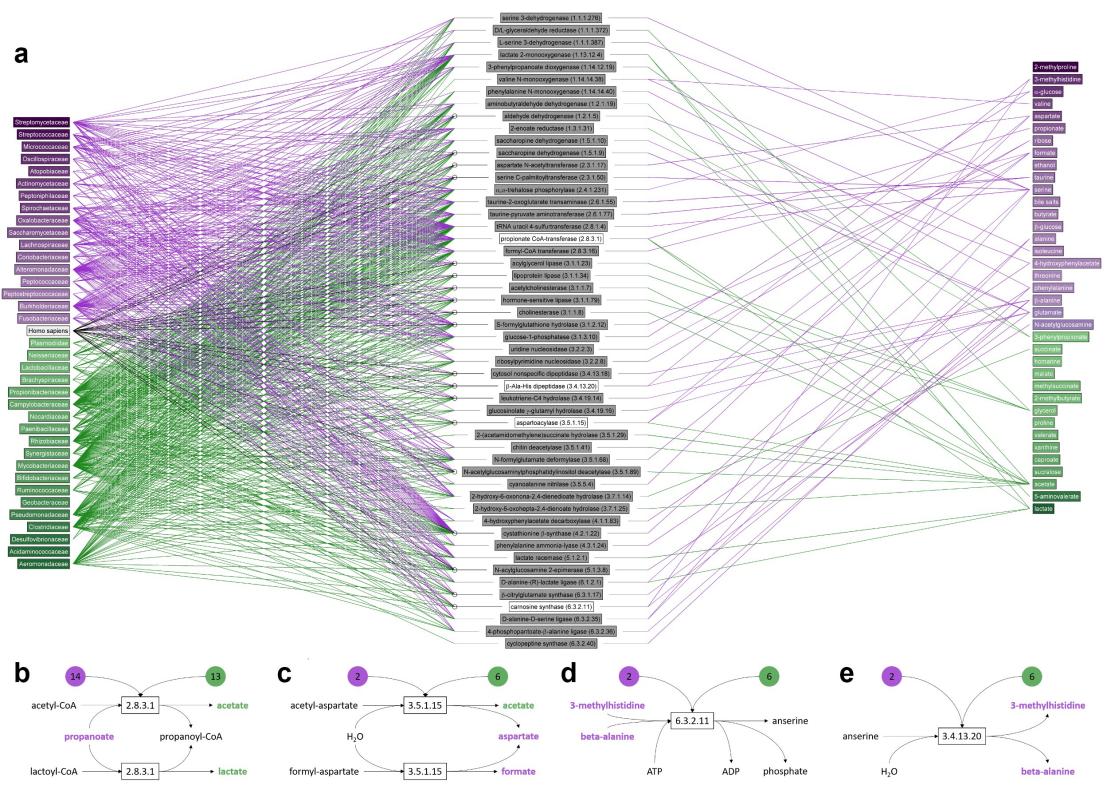 Figure 4 Metabolites involved in reactions mediated by enzymes found in one or more of the microbial families.
Figure 4 Metabolites involved in reactions mediated by enzymes found in one or more of the microbial families.
In this study, the authors used integrated metabolomic and microbiomic analyses to identify fecal microbiome-metabolomic features of IBS that are associated with altered serotonin metabolism and adverse stress responses associated with gastrointestinal symptoms. Metabolic response networks integrated with microbiome information revealed pathways of host-microbiome interactions. These results support a microbiome-gut-brain link in the pathogenesis of IBS.
Reference
- Mujagic, Zlatan, et al. "Integrated fecal microbiome–metabolome signatures reflect stress and serotonin metabolism in irritable bowel syndrome." Gut Microbes 14.1 (2022): 2063016. https://doi.org/10.1080/19490976.2022.2063016
Publications
Here are some publications in Multi-omics Integratives research from our clients:

- Bacterial–fungal interactions revealed by genome-wide analysis of bacterial mutant fitness. 2021. https://doi.org/10.1038/s41564-020-00800-z
- Characterization of a novel AraC/XylS-regulated family of N-acyltransferases in pathogens of the order Enterobacterales. 2020. https://doi.org/10.1371/journal.ppat.1008776
- Insulin resistance does not impair mechanical overload-stimulated glucose uptake, but does alter the metabolic fate of glucose in mouse muscle. 2020. https://doi.org/10.3390/ijms21134715
- Anti-inflammatory activity of black soldier fly oil associated with modulation of tlr signaling: A metabolomic approach. 2023. https://doi.org/10.3390/ijms241310634
- A comprehensive biochemical characterization of settlement stage leptocephalus larvae of bonefish (Albula vulpes). 2021. https://doi.org/10.1111/jfb.14846
References
- Chen, Ziyi, et al. "Integrated metabolome and microbiome strategy reveals the therapeutic effect of nervonic acid on Alzheimer's disease rats." The Journal of Nutritional Biochemistry 137 (2025): 109813. https://doi.org/10.1016/j.jnutbio.2024.109813
- Mannochio-Russo, Helena, et al. "Microbiomes and metabolomes of dominant coral reef primary producers illustrate a potential role for immunolipids in marine symbioses." Communications Biology 6.1 (2023): 896. https://doi.org/10.1038/s42003-023-05230-1







