Exosome Proteomics
Creative Proteomics offers a comprehensive exosome proteomics analysis platform, enabling high-sensitivity identification and quantification of exosome proteins from diverse biological samples. Utilizing advanced mass spectrometry and bioinformatics tools, our platform provides in-depth protein characterization, supporting biomarker discovery and disease researches.
Submit Your Request Now
×- Services
- Workflow
- Advantages
- Applications
- Demo
- FAQs
Exosomes are a type of extracellular vesicle naturally secreted by various cell types, characterized by a diameter ranging from 40 to 160 nm and possessing a bilayer lipid membrane structure. They are derived from outward blebbing of the plasma membrane and widely found in biological fluids, such as peripheral blood, ascites, urine, saliva, cerebrospinal fluid, and other body fluids. Exosomes transport a multitude of bioactive cargoes, including proteins, lipids, nucleic acids (DNA, RNA), and metabolites. They play a crucial role and proteins in exosomes have been revealed to participate in immune regulation, antigen presentation, cancer metastasis, tumor invasion, angiogenesis, and neurodevelopment. Therefore, with the advancement of exosome isolation techniques, there has been a growing interest in elucidating the intricate functions and applications of exosomes in the numerous physiological and pathological processes and hold great potential as early diagnostic markers for various diseases.
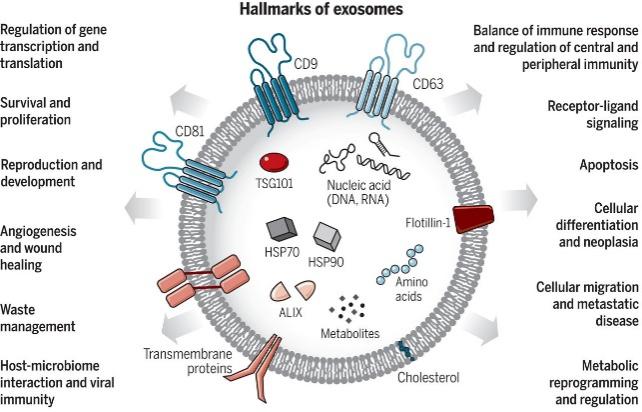 Figure 1. Exosomes: A cell-to-cell transit system in the human body with pleiotropic functions. (Kalluri, R et al., 2020)
Figure 1. Exosomes: A cell-to-cell transit system in the human body with pleiotropic functions. (Kalluri, R et al., 2020)
Exosome Proteomics Service at Creative Proteomics
Creative Proteomics provides comprehensive exosome proteomics analysis services, enabling the identification and quantification of exosome proteins. By integrating bioinformatics analysis, we facilitate biomarker detection through in-depth proteomic data interpretation.
Exosome isolation and purification
Exosome isolation and purification is essential and requires effective and precise extraction. Currently, various methodologies with high specificity and purity have been established. Based on the principle of their separation mechanism, these methods can be divided into three major categories: density, affinity, and size.
Creative Proteomics is capable of conducting diverse exosome sample analyses. The extensive experience we have accumulated over the years enables us to provide a diverse range of exosome isolation and extraction protocols tailored to the specific characteristics of each sample.
The main isolation and purification protocols include:
1. Differential ultracentrifugation method (Current gold standard).
Suitable for large volume samples such as cell supernatant, lavage fluid, urine, etc.
2. Kits based on the principle of PEG precipitation.
Suitable for small volume samples such as plasma, serum, cerebrospinal fluid, etc.
3. Ultrafiltration extraction method.
Suitable for concentration of bacterial cultures or samples with low exosome.
4.Magnetic bead-based extraction method
Exosomes are captured using magnetic nanoparticles with high affinity, followed by washing and elution steps to extract high-purity exosome samples.
Exosome Proteomics Workflow
Exosome Proteomics Workflow involves the isolation, purification, and proteomic analysis of exosomes, essential for disease diagnosis, therapeutic development, and biomarker discovery.
- Exosome isolation and purification aim to obtain high-purity samples using methods like density gradient centrifugation, polymer precipitation, ultrafiltration, and immunomagnetic beads, depending on sample type and research needs.
- Proteomic analysis identifies and quantifies exosome proteins using techniques such as mass spectrometry (LC-MS/MS) with data-dependent (DDA) or data-independent acquisition (DIA) detection mode, 2D-DIGE, and Western blot for validation.
- Data analysis and functional annotation use bioinformatics tools like Proteome Discoverer and MaxQuant for protein identification, PANTHER and UniProt for functional classification, and differential expression analysis to identify biomarkers.
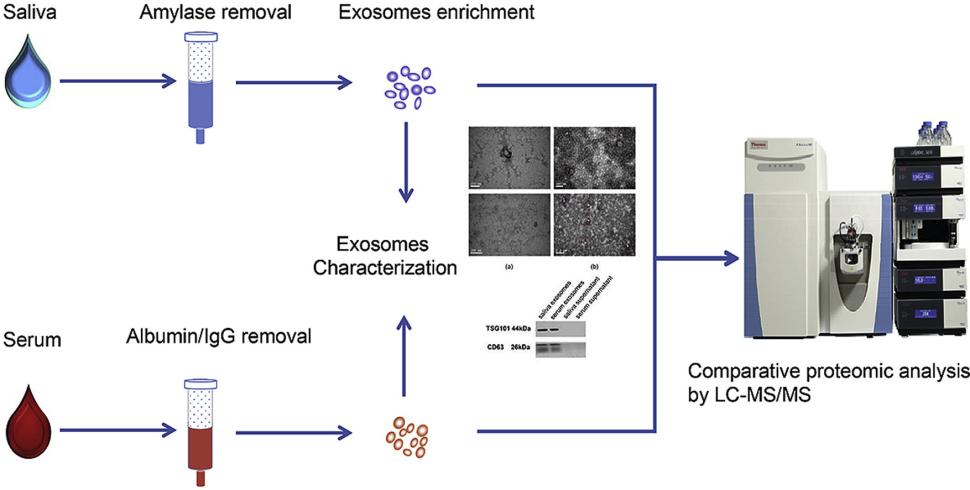 Figure 2. Saliva and serum exosome proteomics workflows. (Y. Sun et al., 2017)
Figure 2. Saliva and serum exosome proteomics workflows. (Y. Sun et al., 2017)
Technical Platforms

Thermo Q ExactiveTM series

AB Sciex 6500+

Thermo Orbitrap Fusion Lumos

Bruker timsTOF Pro
Sample Requirements
| Sample type | Recommended sample size | |
|---|---|---|
| Animal tissues | Hard tissues (bones, hair) | 300-500mg |
| Soft tissues (leaves, flowers of woody plants, herbaceous plants, algae, ferns) | 100mg | |
| Plant tissues | Hard tissues (roots, bark, branches, seeds, etc.) | 3-5g |
| Microbes | Common bacteria, fungal cells (cell pellets) | 100μL |
| cells | Suspension/adherent cultured cells (cell count/pellet) | >1*107 |
| Fluids | Plasma/serum/cerebrospinal fluid | 5 mL |
| Follicular fluid | 200μL | |
| Lymph, synovial fluid, puncture fluid, ascites | 5mL | |
| Saliva/tears/milk | 3-5mL | |
| Others | Culture supernatant (serum-free medium cannot be used) | 20mL |
| Pure protein (best buffer is 8MUrea) | 150μg | |
| Each slice: 10µm thickness, 1.5×2cm area | 15-20 slices | |
| FFPE | ||
Our Advantages
- Professional detection and analysis capability: Experienced technical team, strict quality control system, together with ultra-high resolution detection system and professional data pre-processing and analysis capability, ensure reliable and accurate data.
- Reproducible: Obtain consistent and reproducible inter- and intra- assay results for data analysis.
- High specificity: Established optimized and stabilized exosome isolation and purification methods.
- Multiplex, high-throughput: Deeper coverage of exosome proteins.
- High resolution and sensitivity:, Q-Exactive HF, Orbitrap Fusion™ Tribrid™, Bruker timsTOF Pro2.
Reports
- Detailed report, including experiment procedures, parameters, etc.
- Raw data and data analysis results (database searching results and downstream bioinformatics analysis results, such as volcano plot, GO annotation, PCA, KEGG, Protein Interaction Networks, etc.)
Appliacations of Exosome Proteomics
Early Cancer Diagnosis
Exosome proteomics enables the identification of protein biomarkers for early cancer detection. For example, Glypican-1 (GPC1) on exosomes distinguishes pancreatic cancer patients from healthy individuals.
Tumor Classification and Prognosis
Proteomic analysis of exosomes helps classify cancer types and predict patient prognosis by identifying specific protein markers associated with tumor origin and progression.
Monitoring Disease Progression
Changes in exosomal protein composition over time can be used to track cancer progression and treatment response, offering a non-invasive method for disease monitoring.
Neurodegenerative Disease Biomarkers
Exosomal proteins such as Cathepsin B have been identified as potential biomarkers for Alzheimer's disease, aiding in early diagnosis and understanding disease mechanisms.
Demo Results
(Figures come from Chatterjee, M et al. 2023 )
Exosome Proteomics FAQs
How to isolate and purify exosomes?
There are several current methods of exosome isolation, and it is not possible to specify which isolation method is better; each of them has its own advantages and disadvantages. The separation method needs to be selected according to the characteristics of the sample.
Table 1. Summary of current exosome isolation methods. (Shao, H et al. 2018)
| Method | Mechanism of enrichment | Advantages | Limitations |
|---|---|---|---|
| Ultracentrifugation | Density | 1) Current gold standard; 2) Established protocol. | 1) Lengthy duration (>4 hr); 2) Large sample volume; 3) Requires ultracentrifuge; 4) Low recovery and purity. |
| Sucrose-gradient centrifugation | Density | 1) Current gold standard; 2) High purity. | 1) Lengthy duration (>4 hr); 2) Large sample volume; 3) Requires ultracentrifuge; 4) Low recovery. |
| Co-precipitation | Surface charge | 1) Easy and user-friendly processing. | 1) Lack specificity; 2) Difficulty in scaling. |
| Size-exclusion chromatography | Size and molecular weight | 1) High yield; 2) Wide variety of eluents. | 1) Lack specificity; 2) Difficulty in scaling. |
| Field flow fractionation | Size and molecular weight | 1) Broad separation range; 2) Wide variety of eluents. | 1) Lengthy duration; 2) Requires fractionation equipment. |
| Immunoaffinity enrichment | Affinity | 1) High specificity and purity. | 1) High cost; 2) Low field; 3) Limited use, depending on specificity of the antibody. |
| Microfluidic Filtering | Density and size | 1) Fast and low cost; 2) Convenient and easy to automate. | 1) Low throughout; 2) Complex device. |
| Polymer coprecipitation | Surface charge | 1) Easy and user-friendly processing. | 1) Lacks specificity; 2) difficulty in scaling. |
How to preserve exosomes?
If the study's intention is exosome morphological characteristics, proteins or other functions, it is advisable to utilize freshly isolated exosomes. The current consensus suggests that the optimal utilization of exosomes involves either immediate utilization post-extraction or cryopreservation at -80°C.
How to determine the authenticity of exosomes?
Artificial exosomes are not exosomes.
Lyophilized exosome cultures are not exosomes.
Exosomes may not have strong repair and regeneration ability if they are not derived from stem cells.
Exosomes in liquid form at room temperature do not have biological functional activity and need to be frozen and preserved to maintain its activity.
Learn about other Q&A about other technologies.
Publications
Here are some publications in Proteomics research from our clients:

- Regulation of cardiac ferroptosis in diabetic human heart failure: uncovering molecular pathways and key targets. 2024. https://doi.org/10.1038/s41420-024-02044-w
- Parkinson’s disease-associated LRRK2-G2019S mutant acts through regulation of SERCA activity to control ER stress in astrocytes. 2019. https://doi.org/10.1016/j.yjmcc.2007.03.738
- In Vitro Identification of Phosphorylation Sites on TcPolβ by Protein Kinases TcCK1, TcCK2, TcAUK1, and TcPKC1 and Effect of Phorbol Ester on Activation by TcPKC of TcPolβ in Trypanosoma cruzi Epimastigotes. 2024. https://doi.org/10.3390/microorganisms12050907
- Molecular signature of the ontogenic development of the prawn Macrobrachium tenellum. 2023. https://doi.org/10.7717/peerj.16344
- In vitro assays reveal inherently insecticide-tolerant termite symbionts. 2023. https://doi.org/10.3389/fphys.2023.1134936
References
- Kalluri, R et al. (2020). The biology, function, and biomedical applications of exosomes. Science (New York, N.Y.), 367(6478), eaau6977. https://doi.org/10.1126/science.aau6977
- Sun, Y., et al. (2017). Systematic comparison of exosomal proteomes from human saliva and serum for the detection of lung cancer. Analytica chimica acta, 982, 84–95. https://doi.org/10.1016/j.aca.2017.06.005
- Chatterjee, M et al. (2023). C1q is increased in cerebrospinal fluid-derived extracellular vesicles in Alzheimer's disease: A multi-cohort proteomics and immuno-assay validation study. Alzheimer's & dementia : the journal of the Alzheimer's Association, 19(11), 4828–4840. https://doi.org/10.1002/alz.13066
- Shao, H et al. (2018). New Technologies for Analysis of Extracellular Vesicles. Chemical reviews, 118(4), 1917–1950. https://doi.org/10.1021/acs.chemrev.7b00534


















