Immunopeptidomics Services: Mass Spectrometry-Based Insights for Neoantigen Discovery
Creative Proteomics now offers a cutting-edge immunopeptidomics service that enables high-confidence identification of MHC-bound peptides. By enriching MHC complexes using specific antibodies and analysing their bound peptides on the latest Orbitrap Astral high-resolution mass spectrometer, we provide direct insight into immune-presented peptide sequences—including post-translational modifications often missed by genetic sequencing.
Using de novo sequencing technology, our workflow goes beyond traditional databases to uncover a broader range of MHC-presented peptides, including rare and non-canonical species. Combined with advanced data analysis, this service offers powerful support for neoantigen discovery, immunotherapy development, and next-generation vaccine design.
Submit Your Request Now
×
- High-depth peptide profiling from minimal samples
- Reliable MHC enrichment and spectral quality
- Supports non-canonical and modified peptides
- AI-assisted neoantigen and TCR prediction
- What Is Immunopeptidomics
- Our Services
- FAQ
- Case
- Demo
- Publication
What Is Immunopeptidomics?
Immunopeptidomics is an advanced mass spectrometry technique used to identify and characterise the peptides presented by major histocompatibility complex (MHC) molecules on the surface of cells. These peptides, also called immunopeptides, act as signals that inform the immune system about a cell's internal state.
By mapping these peptide profiles, researchers can uncover what the immune system "sees" in real time — a powerful approach for discovering novel neoantigens, designing personalised cancer vaccines, and investigating the mechanisms behind autoimmune or infectious diseases.
There are two key types of MHC molecules involved:
MHC class I presents peptides from proteins inside the cell (endogenous), mainly interacting with CD8⁺ T cells, which are responsible for killing infected or cancerous cells.
MHC class II, found on specialised antigen-presenting cells, presents peptides derived from both external and internal sources to CD4⁺ T helper cells, which orchestrate broader immune responses.
Why Immunopeptidomics Matters — And Why It's Challenging
T cells rely on immunopeptides to distinguish between healthy cells and foreign threats like viruses or cancer. These short peptides, displayed by MHC molecules on the surface of cells, act as molecular "ID tags" that guide immune responses.
Understanding which peptides are naturally presented — especially those containing neoantigens — is key to designing personalised cancer therapies and uncovering the roots of autoimmune or infectious diseases. Immunopeptidomics provides a direct, high-resolution window into this antigen presentation process by identifying and quantifying MHC-bound peptides.
Why It Matters
- Maps the actual peptide repertoire presented by MHC molecules in different cell states
- Identifies tumour-specific neoantigens for TCR-T therapies and cancer vaccine development
- Detects post-translational modifications (PTMs) relevant to immune recognition
- Supports basic research into autoimmune disorders, chronic infections, and antigen processing defects
Key Challenges
- Complex peptide mixtures: Immunopeptidomics samples contain a vast range of peptides with widely varying abundances
- Low abundance of MHC-bound peptides: Traditional mass spectrometry often struggles to detect rare but clinically relevant peptides
- Diverse peptide sequences: MHC molecules can bind a broad range of peptide lengths and sequences, making identification harder
- Data analysis is demanding: Many peptides are not in standard databases, requiring de novo sequencing and machine learning tools
- Quantitative stability is critical: Accurate and reproducible quantification is essential for biomarker validation and target selection
Our Approach: Integrated Strategies and Advanced Platforms for Immunopeptidomics
At Creative Proteomics, we deliver an end-to-end immunopeptidomics solution tailored to HLA-I peptide analysis. By combining targeted immunopeptide enrichment, cutting-edge mass spectrometry platforms, and intelligent data analytics, we help researchers identify naturally presented peptides with high confidence. This enables accurate neoantigen discovery and supports the development of personalised vaccines, disease models, and immunotherapies.
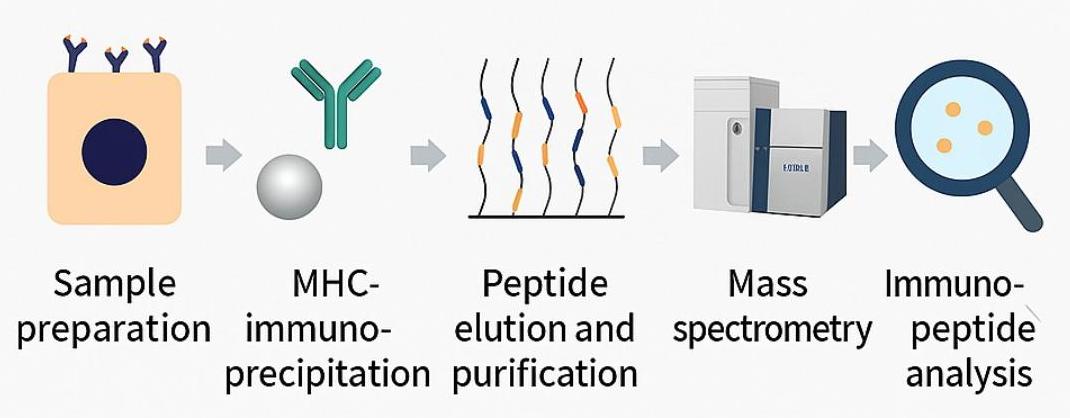
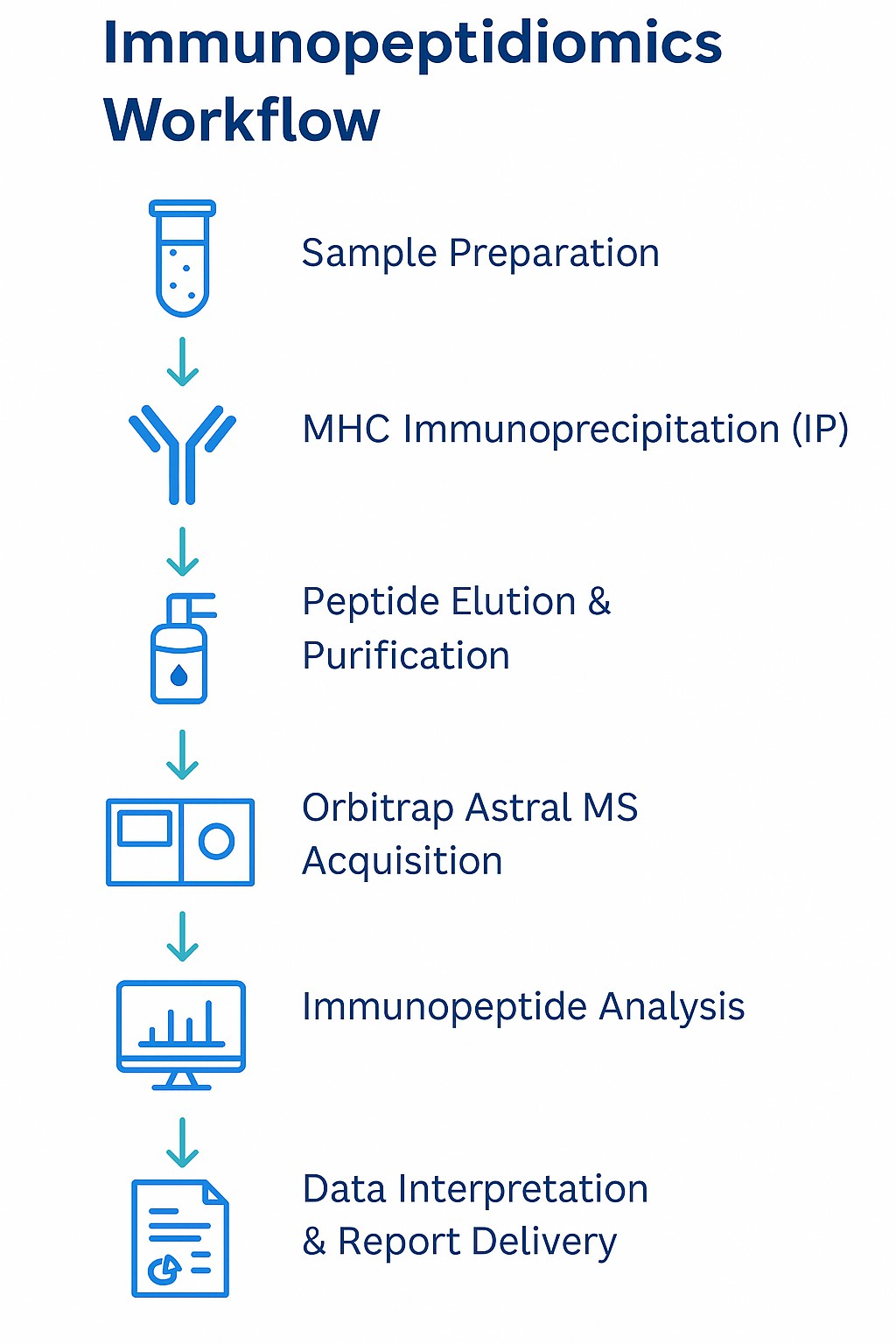
Step-by-Step Workflow: From Sample to Insights
Our immunopeptidomics pipeline integrates seamlessly across six stages:
- Sample Preparation
- MHC Complex Immunoprecipitation (IP)
- Peptide Elution and Purification
- LC-MS/MS Analysis using DDA or DIA acquisition modes
- Peptide Identification via database search + de novo sequencing
- Immunopeptide Annotation and Functional Analysis, including:
- Neoantigen discovery
- TCR recognition prediction
- Post-translational modification mapping
Flexible Acquisition Strategies: DDA vs. DIA
We offer two complementary strategies tailored to different research goals:
▶ DDA (Data-Dependent Acquisition)
- Generates clean, interpretable spectra, ideal for de novo sequencing
- Excellent for identifying non-canonical peptides and novel neoepitopes
- Works best in discovery-driven studies such as personalised vaccine development
- Fully compatible with DeepNovo® for high-precision peptide decoding
▶ DIA (Data-Independent Acquisition)
- Offers broader coverage and improved repeatability, especially for low-abundance samples
- Enables high-throughput quantification with exceptional stability
- Ideal for targeted antigen verification, large-scale profiling, and quantitative biomarker discovery
DDA vs DIA: How to Choose?
| Feature | DDA Strategy | DIA Strategy |
|---|---|---|
| Best for | Neoantigen discovery, rare variant detection, de novo analysis | Target quantification, biomarker studies, low-input samples |
| Peptide ID Depth | High, especially for novel peptides | Even higher (+7% yield), excels at low-input |
| Reproducibility | Moderate | High — up to 74.5% overlap across replicates |
| Quantification | ~73% of peptides quantifiable | ~100% quantifiable, 94% with CV < 20% |
| Recommended Use | Personalised cancer vaccines, mutation studies | Antigen validation, TCR-T workflows, TMG assays |
AI-Powered Analysis
To maximise the precision and depth of immunopeptidomics results, Creative Proteomics integrates a suite of AI platforms into every stage of data interpretation:
- A proprietary platform that combines mass spectrometry data with advanced machine learning to support neoantigen discovery, immunogenicity scoring, and TCR binding prediction.
- One of the AI-driven pipelines designed specifically for immunopeptides, enabling accurate identification of non-canonical peptides that are not captured by standard databases.
- A machine learning-enhanced de novo sequencing tool, fine-tuned for HLA-presented peptides, improving sequence confidence and detection of rare variants.
Advanced Instrumentation and Analytical Strength
| Platform/Tool | Key Advantages |
|---|---|
| Orbitrap Astral | Ultra-high resolution and sensitivity for reliable detection in low-input samples |
| nanoElute® 2 System | Exceptional system stability and reproducibility, ideal for micro-scale workflows |
| Integrated Data Hub | Supports de novo sequencing, PTM detection, TCR mapping, and immunogenicity scoring |
What Sets Our Service Apart: Four Dimensions of Performance
Comprehensive
- Identify 30,000+ peptides in a single experiment, covering both MHC-I and MHC-II pathways
- Maintain high performance with low-input samples — detect over 5,000 peptides from just 10 mg tissue or 20 million cells
- Reveal non-canonical peptides (e.g., frameshift, fusion, cryptic peptides) without database reliance
- Profile more than 10,000 unique HLA-I and HLA-II peptides, greatly expanding the neoantigen discovery landscape
Efficient
- One-stop solution from sample prep to TCR prediction and visualised reporting
- Modular options available: peptide identification, HLA typing, immunogenicity scoring, or antigen filtering
- Deliverables include peptide tables, HLA-binding affinity data, motif distribution, PTM annotation, and top-ranking neoantigen candidates
Reliable
- Powered by nanoElute® 2 + timsTOF Pro 2 with PASEF® — delivers ultra-deep peptide identification with minimal input
- DIA mode ensures exceptional consistency: >74% peptide overlap across replicates, and 94% with CV<20%
- Full-chain quality control tracks peptide coverage, enrichment efficiency, spectral quality, and quantification precision
- Operated by an expert team following rigorous SOPs to ensure reproducibility, traceability, and accuracy
Precise
- AI-guided analytics identify high-confidence immunogenic peptides with or without genome sequencing support
- Multilayer scoring models assess HLA-binding affinity, TCR recognition likelihood, motif conservation, and PTM impact
- Detects PTMs that are often missed by NGS-based workflows, such as phosphorylation or acetylation
- Supports prioritisation of immunotherapy targets most likely to elicit T cell responses
Sample Submission Guidelines
| Sample Type | Project Type | Minimum Amount Required |
|---|---|---|
| Tissue (human or mouse) | MHC-I / MHC-II | ≥ 50 mg per sample |
| Cell lines (human/mouse) | MHC-I / MHC-II | ≥ 2 × 10⁷ cells per sample |
| PBMC (human or mouse) | MHC-I / MHC-II | ≥ 2 × 10⁷ cells per sample |
| Serum or plasma (human) | MHC-II only | ≥ 5 mL per sample (HLA-I not supported) |
Note: One sample can be analysed for both MHC-I and MHC-II. For other species or sample types, please consult our scientific advisors.
Application
| Application Area | Study Focus | Source |
|---|---|---|
| Target Discovery & Immunotherapy | Neoantigen screening in diffuse intrinsic pontine glioma | A combined immunopeptidomics, proteomics, and cell surface proteomics approach to identify immunotherapy targets for diffuse intrinsic pontine glioma, 2023 |
| HLA-A*11:01-restricted neoepitope identification | Identification of neoepitope reactive T-cell receptors guided by HLA-A*03:01 and HLA-A*11:01 immunopeptidomics, 2023 | |
| Instability of HIV antigen presentation | Instability of the HLA-E peptidome of HIV presents a major barrier to therapeutic targeting, 2024 | |
| Basic Research & Mechanism Studies | HPV E5-mediated immune evasion via antigen processing disruption | Human papillomavirus E5 suppresses immunity via inhibition of the immunoproteasome and STING pathway, 2023 |
| MHC-I–driven CD8⁺ T cell activation in NASH | Myeloid cell MHC I expression drives CD8+ T cell activation in nonalcoholic steatohepatitis, 2024 |
Deliverary
- Identified immunopeptide list (sequence, modifications, abundance)
- MHC binding site prediction and motif analysis
- Ranked neoantigen candidates
- TCR recognition potential
- De novo–identified non-canonical peptides
- Visual outputs: heatmaps, sequence logos, distribution plots
All results are provided in both raw and processed formats.
FAQ
What distinguishes your immunopeptidomics service from other peptide identification methods?
Our service directly identifies peptides presented by MHC molecules using high-resolution mass spectrometry. This approach captures naturally processed peptides, including non-canonical and post-translationally modified variants, offering insights that genomic or transcriptomic analyses might miss.
Can this service aid in neoantigen discovery for immunotherapy development?
Absolutely. By profiling the immunopeptidome, we can identify tumor-specific neoantigens, which are crucial for designing personalized cancer vaccines and T-cell receptor (TCR) therapies.
What types of samples are suitable for immunopeptidomics analysis?
We accept a variety of sample types, including cell lines, tissue biopsies, and peripheral blood mononuclear cells (PBMCs). Our protocols are optimized to handle samples with varying cell counts and tissue amounts.
How do you ensure the reliability and reproducibility of the results?
We employ stringent quality control measures throughout the workflow, from sample preparation to data analysis. Our use of advanced instrumentation and standardized protocols ensures high reproducibility and accuracy.
Is it possible to detect low-abundance peptides using your platform?
Yes. Our sensitive mass spectrometry techniques are capable of detecting low-abundance peptides, which is essential for comprehensive immunopeptidome profiling.
Do you provide bioinformatics support for data interpretation?
Yes. Our bioinformatics team offers comprehensive analysis, including peptide identification, MHC binding predictions, and neoantigen ranking, to facilitate downstream applications.
How customizable is your service to specific research needs?
Our service is highly flexible. We can tailor the workflow to focus on specific MHC classes, incorporate particular enrichment strategies, or adjust analytical parameters to meet unique project requirements.
What deliverables can I expect from this service?
Clients receive a detailed report encompassing identified peptides with annotations, MHC binding predictions, neoantigen candidates, and visual data representations such as heatmaps and sequence logos.
How do I initiate a project or get a consultation?
You can contact our team through our website or email to discuss your project needs. We offer consultations to design a workflow that aligns with your research objectives.
Learn about other Q&A.
Client Case Study: Immunopeptidomics Reveals How Mosquito Saliva Modulates Human Immunity to Facilitate Viral Infection

DOI: 10.1126/sciimmunol.adk9872
- Background
- What We Provided
- Results & Value
In a landmark study published in Science Immunology (2024), researchers from Yale University discovered that a salivary protein from Aedes aegypti, named Nest1, directly interacts with the human immune checkpoint CD47. This interaction suppresses skin-resident immune responses, thereby enhancing the infectivity of arboviruses like Zika at the bite site.
Creative Proteomics played a key role in this discovery by delivering a suite of targeted immunoproteomic services, including:
- High-Resolution Mass Spectrometry (MS) for Protein Identification
→ We performed deep proteomic profiling of mosquito saliva to help identify the novel Nest1 protein and assess its abundance and structure. - MHC-Associated Peptide Analysis via Immunopeptidomics
→ Our MHC-peptide enrichment and LC-MS/MS pipeline enabled the detection of host-derived peptides presented in the skin microenvironment, helping clarify how Nest1 modulates antigen presentation pathways. - PTM Mapping and Peptide Annotation
→ Using proprietary bioinformatics tools, we annotated post-translational modifications on key immune components affected by Nest1, illuminating mechanisms of immune suppression. - Data Interpretation Support
→ We provided downstream analysis to support conclusions on CD47 pathway disruption and T cell response inhibition, accelerating insight into host-pathogen interaction.
Our services enabled the researchers to:
- Unambiguously identify Nest1 as an immunosuppressive effector.
- Validate its interaction with CD47 via proteomics-informed pathway analysis.
- Reveal downstream immune consequences using peptide presentation data.
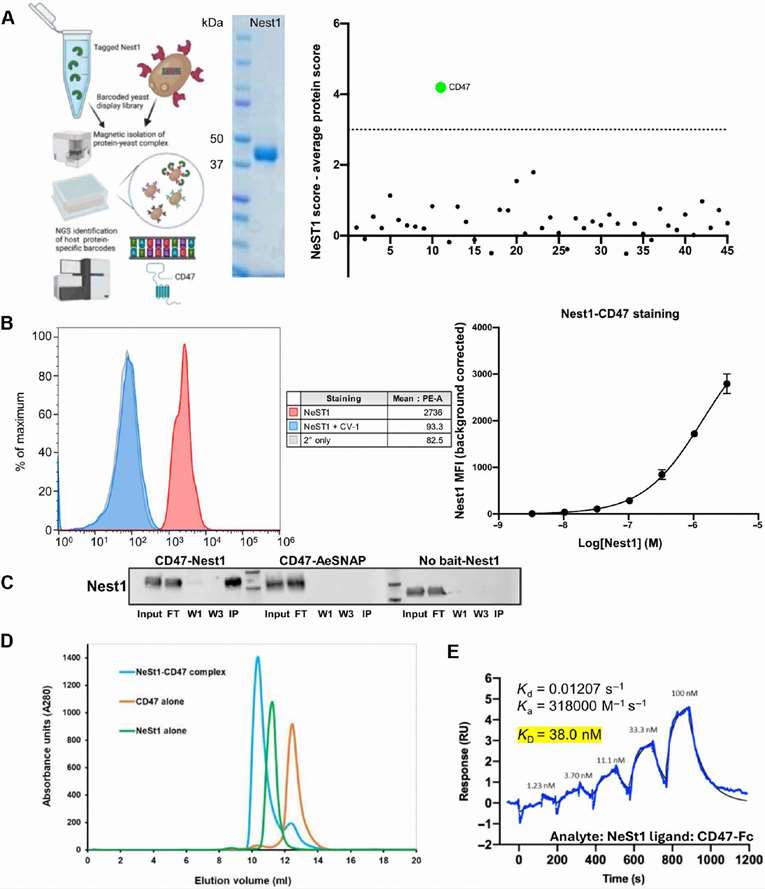 Characterization of the Nest1-CD47 interaction.
Characterization of the Nest1-CD47 interaction.
This insight is crucial for understanding vector-borne disease enhancement and could inform vaccine development targeting saliva-derived immunomodulators.
Demo
Publication

- The human CD47 checkpoint is targeted by an immunosuppressive Aedes aegypti salivary factor to enhance arboviral skin infectivity. Science immunology. 2024. DOI: 10.1126/sciimmunol.adk9872
- Immunoproteomics characterization of Leishmania panamensis proteins for potential clinical diagnosis of mucosal Leishmaniasis. Parasite Immunology. 2021. https://doi.org/10.1111/pim.12824
- Conformational differences between functional human immunodeficiency virus envelope glycoprotein trimers and stabilized soluble trimers. Journal of Virology. 2019. https://doi.org/10.1128/jvi.01709-18
References
- Xie N,Shen G,Gao W, et al. Neoantigens: promising targets for cancer therapy. Signal Transduct Target Ther. 2023;8 (1):9. https://doi.org/10.1038/s41392-022-01270-x
- Blunt MD,Fisher H,Schittenhelm RB, et al. The nuclear export protein XPO1 provides a peptide ligand for natural killer cells. Sci Adv. 2024;10 (34):eado6566. doi:10.1126/sciadv.ado6566
- Chiaro J,Antignani G,Feola S, et al. Development of mesothelioma-specific oncolytic immunotherapy enabled by immunopeptidomics of murine and human mesothelioma tumors. Nat Commun. 2023;14 (1):7056. https://doi.org/10.1038/s41467-023-42668-7
- Mayer RL,Verbeke R,Asselman C, et al. Immunopeptidomics-based design of mRNA vaccine formulations against Listeria monocytogenes. Nat Commun. 2022;13 (1):6075. https://doi.org/10.1038/s41467-022-33721-y













