Sphingolipid Metabolism Analysis Service
Sphingolipid metabolism is tightly linked to cellular function, signaling, and membrane dynamics. Creative Proteomics provides targeted and discovery-based sphingolipidomics service empowers you to decode these biochemical layers with precision LC-MS/MS and Orbitrap technologies.
✓ Absolute quantification of 200+ sphingolipids
✓ Advanced targeted & untargeted MS workflows
✓ Compatible with biofluids, cells, tissues, and organelles
✓ Optimized extraction and chromatographic resolution
✓ Detailed reports with pathway insights and multivariate analysis
Submit Your Request Now
×
What You Will Receive:
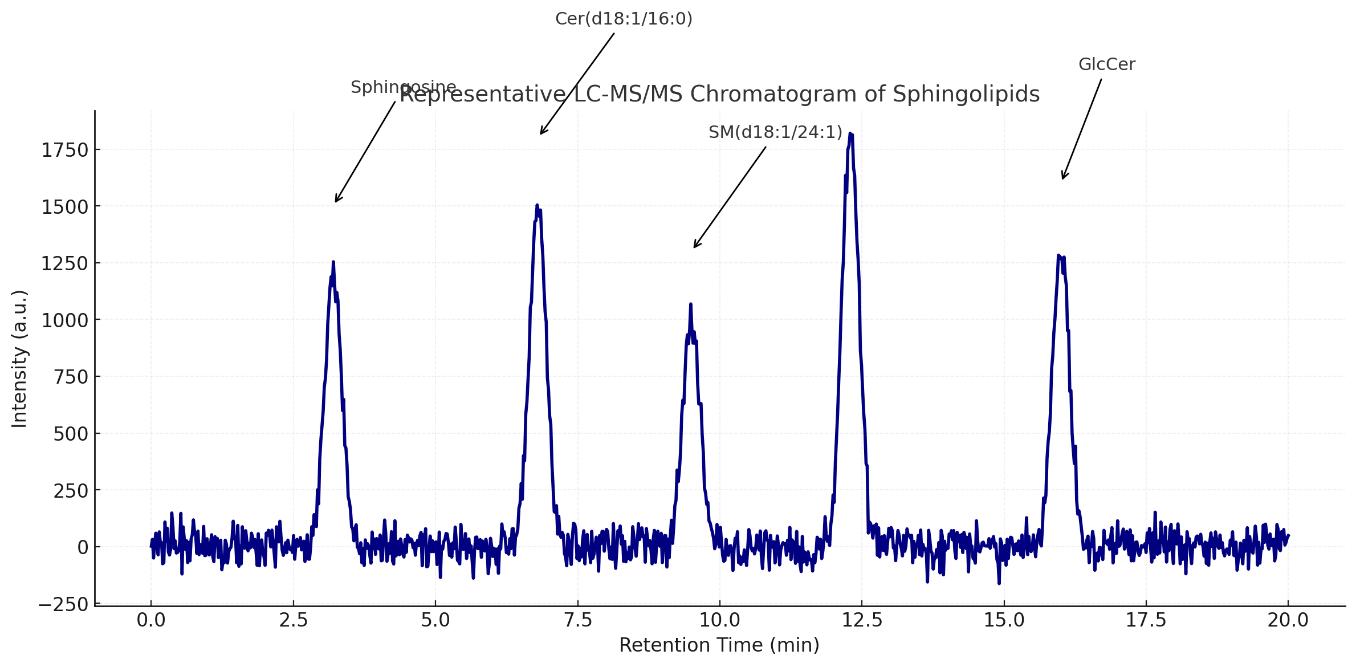
- High-resolution chromatograms
- Absolute and relative lipid quantification
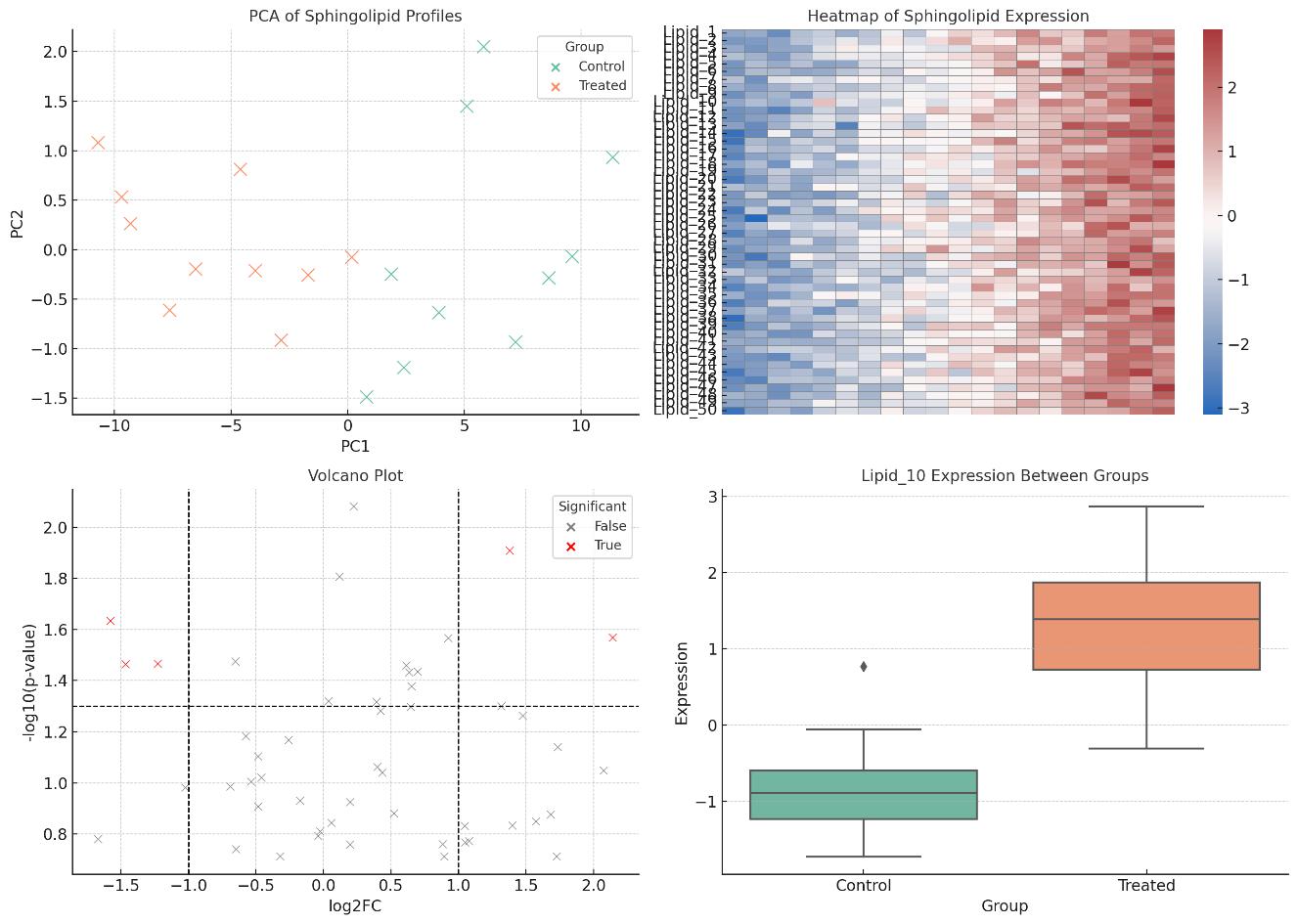
- Multivariate analysis: PCA, heatmaps, volcano plots
- QC metrics and reproducibility report
- Optional: Biological annotations & pathway visualization
- What We Provide
- Advantages
- Technology Platform
- Sample Requirement
- Demo
- FAQ
What Are Sphingolipids?
Sphingolipids are a structurally diverse class of lipids integral to membrane architecture and cellular signaling. Unlike glycerophospholipids, sphingolipids are derived from a sphingoid base backbone and include key species such as ceramides, sphingomyelins, glycosphingolipids, and sphingosine-1-phosphate. These lipids are tightly regulated through intricate metabolic pathways and play critical roles in modulating cell growth, differentiation, apoptosis, immune responses, and neurodegeneration.
Even subtle dysregulation in sphingolipid metabolism can trigger major pathophysiological consequences. Thus, accurate and high-resolution sphingolipid profiling is essential for biomedical research, drug development, and functional lipidomics.
Why You Need Sphingolipid Metabolism Analysis
Targeted and high-throughput sphingolipid analysis empowers researchers and developers to:
- Identify and quantify specific lipid species within complex biological matrices
- Reveal metabolic imbalances that underlie diseases such as cancer, metabolic syndromes, and neurodegenerative conditions
- Monitor biomarker candidates in preclinical studies
- Evaluate the efficacy and mechanism-of-action of lipid-modulating therapeutics
- Enable comprehensive systems biology interpretations by integrating lipidomic data with transcriptomic and proteomic findings
At Creative Proteomics, we align our service design with your research objectives—whether you need to unravel subtle metabolic shifts or conduct robust lipid screening in large-scale studies.
Our Service Portfolio: Creative Proteomics's Sphingolipids Analysis
- Quantitative Sphingolipid Profiling: Accurate measurement of key sphingolipid species using internal standards for absolute quantification.
- Differential Sphingolipid Expression Analysis: Compare sphingolipid levels across experimental conditions to identify biological changes and potential biomarkers.
- Subcellular Sphingolipid Analysis: Profile sphingolipid distribution within isolated organelles (e.g., mitochondria, ER) for insights into compartment-specific metabolism.
- Sphingolipid Pathway Flux Analysis: Use stable isotope-labeled substrates to track dynamic changes and metabolic turnover in sphingolipid pathways.
- Custom Sphingolipid Panel Development: Design targeted panels based on research needs, including rare or modified sphingolipid species and pathway-specific targets.
- Matrix-Compatible Sample Handling: Validated methods for various sample types including blood, CSF, urine, feces, and cultured cells to ensure reliable lipid recovery.
- Data Analysis & Biological Interpretation: Comprehensive data reporting with multivariate statistical analysis and biological pathway insights included.
List of Detected Sphingolipids and Related Metabolites
| Class | Representative Analytes | Notes |
|---|---|---|
| Ceramides (Cer) | C14:0-Cer, C16:0-Cer, C18:0-Cer, C20:0-Cer, C22:0-Cer, C24:0-Cer, C24:1-Cer, C26:0-Cer | N-acylated sphingosines; core intermediates |
| Dihydroceramides (dhCer) | C14:0-dhCer, C16:0-dhCer, C18:0-dhCer, C22:0-dhCer, C24:0-dhCer, C24:1-dhCer | Saturated analogs of ceramides |
| Sphingomyelins (SM) | SM(d18:1/16:0), SM(d18:1/18:0), SM(d18:1/20:0), SM(d18:1/22:0), SM(d18:1/24:0), SM(d18:1/24:1) | Major membrane lipids, abundant in plasma |
| Sphingosines | Sphingosine (d18:1), Sphinganine (d18:0), Phytosphingosine (t18:0) | Free bases; key signaling molecules |
| Phosphorylated Sphingoid Bases | Sphingosine-1-phosphate (S1P), Sphinganine-1-phosphate (dhS1P) | Potent bioactive lipids; immune and vascular roles |
| Ceramide-1-phosphate (C1P) | C16:0-C1P, C18:0-C1P, C24:1-C1P, C24:0-C1P | Phosphorylated ceramide; inflammatory mediator |
| Glycosylceramides (HexCer) | Glucosylceramide (GlcCer), Galactosylceramide (GalCer), LacCer | Important in lysosomal storage and signaling |
| Gangliosides | GM3, GM2, GM1, GD1a, GD1b, GT1b | Sialic acid-containing glycosphingolipids; abundant in brain |
| 3-Ketosphinganine | 3-Ketosphinganine | Precursor in de novo sphingolipid synthesis |
| Dehydrosphinganine | Transient biosynthetic intermediate | Often included in extended pathway panels |
| Other Atypical Sphingolipids | 1-Deoxysphinganine, 1-Deoxymethylsphinganine | Associated with metabolic disorders and toxicity |
Advantages of Sphingolipids Assay
- High Sensitivity: Detection limits as low as 0.05 ng/mL for key sphingolipid species (e.g., sphingosine-1-phosphate, ceramides), enabling analysis of low-abundance lipids in biofluids and tissue samples.
- Broad Coverage: Capable of quantifying over 200 sphingolipid species across major classes including ceramides, dihydroceramides, sphingomyelins, glycosphingolipids, and sphingosine phosphates.
- Excellent Reproducibility: Intra- and inter-batch CVs<10% for most analytes; strict QC monitoring ensures consistent performance across large-scale studies.
- Wide Dynamic Range: Linear quantification across four to five orders of magnitude, suitable for samples with both trace-level and abundant sphingolipids.
- Flexible Sample Compatibility: Validated extraction protocols for serum, plasma, CSF, urine, cultured cells, tissue homogenates, and other complex biological matrices.
- Optimized Chromatographic Separation: Tailored UHPLC gradients provide baseline resolution of isobaric sphingolipid species, ensuring confident identification and accurate quantification.
- Comprehensive Data Reporting: Includes absolute concentrations (ng/mL or pmol/mg), QC summaries, MRM transitions, retention time tables, and optional statistical plots (e.g., PCA, heatmaps, volcano plots).
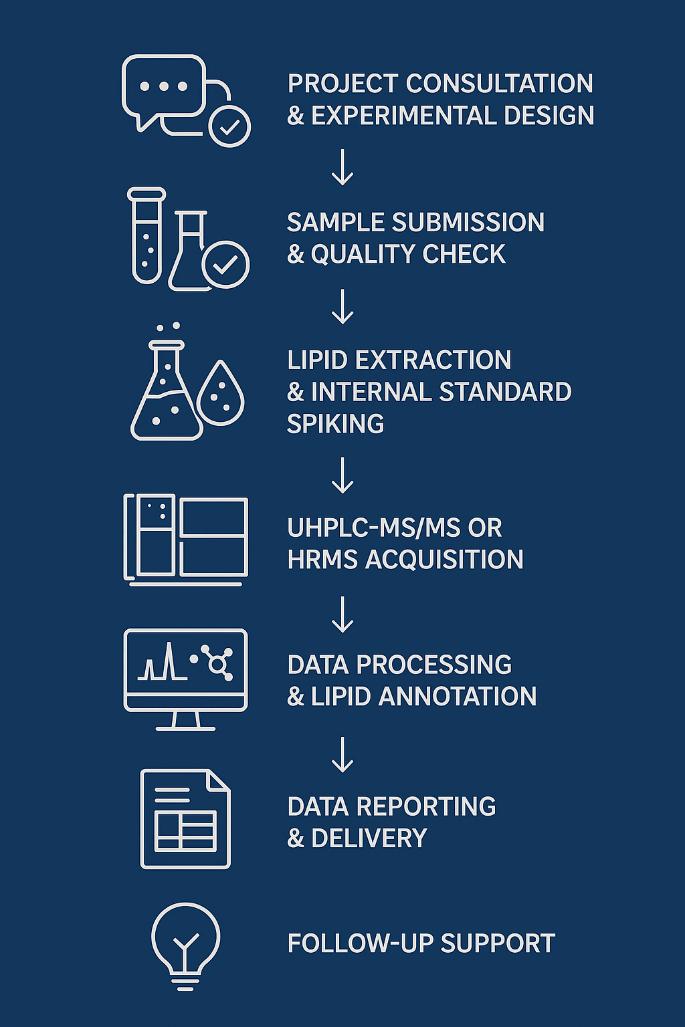
Workflow for Sphingolipids Analysis Service
1. Project Consultation & Experimental Design
Discuss study goals, sample types, target sphingolipids, and analytical needs. Recommend appropriate method (targeted vs. untargeted), platform, and detection scope.
2. Sample Submission & Quality Check
Acceptable matrices: plasma, serum, tissue, CSF, cultured cells, etc. Perform visual inspection, volume verification, and pre-processing QC as needed.
3. Lipid Extraction & Internal Standard Spiking
Use of optimized extraction protocols (e.g., MTBE or Bligh & Dyer) to isolate sphingolipids. Spike with stable isotope-labeled internal standards for accurate quantification.
4. UHPLC-MS/MS or HRMS Acquisition
- Targeted quantification via LC-6495C Triple Quad MS/MS (MRM mode).
- Untargeted profiling via Orbitrap Exploris HRMS (Full MS/DDA/DIA).
- Chromatographic separation on C18 UHPLC column ensures peak resolution.
5. Data Processing & Lipid Annotation
Use software tools such as MultiQuant™, LipidSearch™, or Skyline for peak integration. Structural confirmation of lipid species, isomers, and phosphorylated forms.
6. Data Reporting & Delivery
Deliver raw data files, processed concentration tables, QC reports, and summary plots. Optional statistical analysis (e.g., PCA, heatmaps) available upon request.
Technology Platform for Sphingolipids Analysis Service
LC-MS/MS Platform
- Instrument: Agilent 6495C Triple Quadrupole LC/MS System
- Ionization Mode: Positive Electrospray Ionization (ESI+)
- Operating Mode: Multiple Reaction Monitoring (MRM)
- Dynamic Range: Up to 10⁴ fold (from ~0.05 ng/mL to >1,000 ng/mL)
- Typical LOD: As low as 0.05 ng/mL for ceramides and sphingosine-1-phosphate, depending on matrix and chain length
 Agilent 6495C Triple Quadrupole (Figure from Agilent)
Agilent 6495C Triple Quadrupole (Figure from Agilent)
UHPLC Separation
- UHPLC System: Agilent 1290 Infinity II or Thermo Scientific™ Vanquish™ Flex UHPLC
- Column: C18 column, 1.7 µm particle size, 2.1 × 100 mm
- Flow Rate: 0.3 mL/min
- Column Temperature: 45°C
- Mobile Phases:
A: Water + 0.1% Formic Acid
B: Acetonitrile/Isopropanol (1:1) + 0.1% Formic Acid - Gradient Program:
Tailored gradient profile optimized for baseline separation of ceramides, dihydroceramides, sphingomyelins, glycosphingolipids, and sphingosine derivatives with high peak resolution and retention stability.
 Agilent 1260 Infinity II HPLC (Fig from Agilent)
Agilent 1260 Infinity II HPLC (Fig from Agilent)
High-Resolution MS Platform
- Instrument: Thermo Scientific™ Orbitrap Exploris™ 240/480 Mass Spectrometer
- Ionization Mode: Positive & Negative Electrospray Ionization (ESI±)
- Operating Mode: Full MS, Data-Dependent MS/MS (DDA), and Data-Independent Acquisition (DIA)
- Mass Resolution: Up to 480,000 FWHM at m/z 200
- Mass Accuracy:<2 ppm with internal calibration
- Typical LOD: Low femtomole range for sphingolipids depending on compound class
This platform provides exceptional resolution and mass accuracy for untargeted sphingolipidomics, enabling confident identification of isobaric lipids, complex glycosphingolipids, and novel sphingoid derivatives. Ideal for discovery-based and structural elucidation workflows.
 Orbitrap Exploris™ 240/480 Mass Spectrometer
Orbitrap Exploris™ 240/480 Mass Spectrometer
(Fig from Thermo Scientific)
Sample Requirements for Sphingolipids Analysis Service
| Sample Type | Recommended Volume / Amount | Storage Conditions | Transport Requirements |
|---|---|---|---|
| Plasma / Serum | ≥ 100 µL | -80°C | Dry ice shipment; avoid freeze-thaw cycles |
| Tissue | ≥ 20 mg (wet weight) | Snap frozen in liquid N₂, then -80°C | Ship on dry ice; no preservatives |
| Cultured Cells | ≥ 1 × 10⁶ cells (cell pellet) | -80°C | Ship cell pellet on dry ice |
| CSF (Cerebrospinal Fluid) | ≥ 100 µL | -80°C | Ship on dry ice; pre-filter if necessary |
| Urine | ≥ 1 mL (concentrated if needed) | -80°C | Ship frozen; no additives |
| Fecal Samples | ≥ 200 mg (fresh weight) | -80°C | Snap freeze and ship on dry ice |
| Lysates / Extracts | ≥ 50 µL or equivalent | -80°C | Freeze immediately; ship on dry ice |
Demo Results
FAQ of Sphingolipids Analysis Service
Can this assay distinguish between isomeric or closely related sphingolipid species?
Yes. Our platform uses high-resolution MS and optimized UHPLC separation, enabling clear resolution of isobaric sphingolipids such as Cer(d18:1/24:0) vs. Cer(d18:1/24:1) and regioisomers of sphingosine phosphates.
Is the method suitable for both targeted and untargeted sphingolipid analysis?
Yes. We provide both targeted quantification using triple quadrupole LC-MS/MS and untargeted profiling via Orbitrap-based HRMS, depending on the project's research goals.
Can you analyze glycosphingolipids and complex derivatives?
Absolutely. Our method library includes gangliosides (GM1, GM3), globosides, and other glycosylated ceramides, with optimized fragmentation strategies to aid structural elucidation.
How is data quality and reproducibility ensured?
We implement strict QC workflows, including internal standards, pooled QC samples, and system suitability checks. RSD values for key targets are typically below 10% across replicates.
Are stable isotope-labeled internal standards used for quantification?
Yes. Wherever available, we use isotope-labeled standards to improve accuracy and correct for matrix effects and instrument variability.
Can your method support pharmacokinetic or dose-response studies?
Definitely. Our platform supports sensitive and linear quantification over a wide dynamic range, making it suitable for kinetic studies and compound efficacy evaluation.
How are sphingolipid subclasses differentiated during analysis?
We employ subclass-specific MRM transitions and retention time windows for precise annotation of ceramides, sphingomyelins, hexosylceramides, lactosylceramides, and complex gangliosides. HRMS with MS/MS fragmentation is used to resolve overlapping backbones and headgroups.
Is lipid extraction performed using a standardized method compatible with sphingolipid chemistry?
Yes. We use a modified Bligh-Dyer or MTBE-based extraction tailored to retain amphipathic sphingolipids with minimal degradation or loss. Internal standards are spiked before extraction to account for process variability.
Can this platform quantify both endogenous and exogenous sphingolipid species?
Yes. Our assay can distinguish between labeled (e.g., ^13C/^15N) and endogenous species, supporting metabolic flux studies and compound tracking experiments.
How are structural isomers confirmed in untargeted datasets?
Structural isomers are validated by tandem MS fragmentation spectra (MS²), retention time libraries, and when necessary, authentic standards or ion mobility-assisted resolution.
Are the methods compatible with downstream statistical or network analysis tools?
Yes. We deliver results in publication-ready formats (CSV, Excel, and annotated plots), fully compatible with MetaboAnalyst, Cytoscape, SIMCA, or R/Python-based statistical pipelines.
What is the minimum sample volume required for low-abundance sphingolipids?
Depending on the matrix, as little as 20–30 µL of plasma or 10 mg of tissue may be sufficient, owing to the assay's low LODs and high sensitivity (down to low femtomole levels).
Can pathway-level interpretation be integrated into the report?
Yes. Upon request, we provide pathway mapping to KEGG/LIPID MAPS sphingolipid pathways, enrichment analysis, and visualization of class-level trends (e.g., Cer/SM ratios, salvage vs. de novo synthesis flux).
How are batch effects and instrument drift controlled?
We incorporate pooled QC injections every 8–10 runs, monitor internal standard recovery, and apply signal correction algorithms (e.g., LOESS, PQN) to normalize across batches.
Learn about other Q&A about proteomics technology.
Publications
Here are some of lipidomics-related papers published by our clients:

- White matter lipid alterations during aging in the rhesus monkey brain. 2024. https://doi.org/10.1007/s11357-024-01353-3
- Characterization of Dnajc12 knockout mice, a model of hypodopaminergia. 2024. https://doi.org/10.1101/2024.07.06.602343
- Evidence for phosphate-dependent control of symbiont cell division in the model anemone Exaiptasia diaphana. 2024. https://doi.org/10.1128/mbio.01059-24
- The olfactory receptor Olfr78 promotes differentiation of enterochromaffin cells in the mouse colon. 2024. https://doi.org/10.1038/s44319-023-00013-5
- Annexin A2 modulates phospholipid membrane composition upstream of Arp2 to control angiogenic sprout initiation. 2023. https://doi.org/10.1096/fj.202201088R
- Lipid Membrane Engineering for Biotechnology. 2023. https://doi.org/10.48780/publications.aston.ac.uk.00046663