Sphingoid Bases Profiling Service
Every cell speaks in lipids, and sphingoid bases hold some of its most critical secrets. From ensuring product quality in pharma or cosmetics to exploring cellular pathways, precise analysis of these molecules makes the difference between assumptions and answers. Creative Proteomics gives you the data clarity to move forward with confidence.
- Detect trace levels down to 0.05 ng/mL
- Tailored assays for biological and industrial samples
- Clear differentiation of free vs. phosphorylated forms
- Detailed reports with quantitative insights
- Scalable solutions for research and QC
Submit Your Request Now
×
What You Will Receive:
- Quantitative results for sphingoid bases and derivatives
- High-resolution chromatograms
- Calibration curves and QC data
- Expert interpretation tailored to your research or product goals
- What We Provide
- Advantages
- Technology Platform
- Sample Requirement
- Demo
- FAQ
What Are Sphingoid Bases?
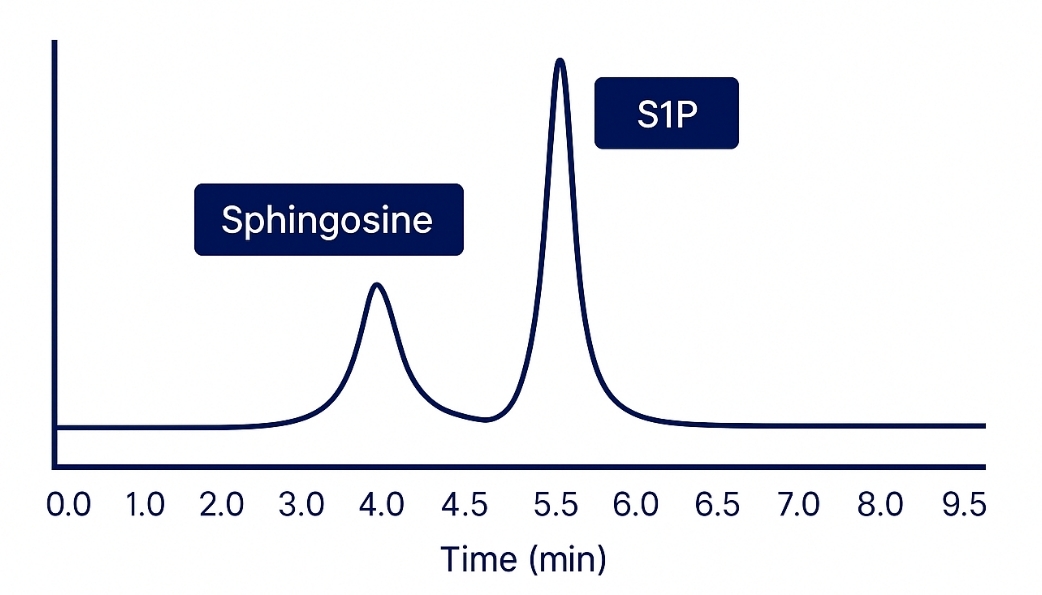
Sphingoid bases are long-chain amino alcohols that serve as the structural backbone of sphingolipids—essential biomolecules integral to cellular membranes, signaling pathways, and lipid metabolism. Major sphingoid bases include sphingosine, dihydrosphingosine (sphinganine), and phytosphingosine, each differing by degree of unsaturation or hydroxylation. Their phosphorylated counterparts, such as sphingosine-1-phosphate (S1P), act as potent signaling molecules implicated in cell proliferation, apoptosis, inflammation, and metabolic regulation.
In biological systems and industrial bioprocesses, fluctuations in sphingoid base profiles are increasingly scrutinized for their roles in:
- Cell membrane architecture and dynamics
- Lipid rafts and signaling platforms
- Stress responses and cell survival mechanisms
- Quality assessment in pharmaceutical, food, and cosmetic applications
- Metabolic pathway elucidation in biotechnology research
Why Analyze Sphingoid Bases?
Analyzing sphingoid bases yields critical insights for both research and industrial sectors. Precise quantification helps:
- Characterize Metabolic Pathways: Understand sphingolipid biosynthesis and catabolism for functional studies or strain engineering.
- Assess Product Quality & Safety: Monitor bioactive lipid levels in pharmaceutical and cosmetic formulations to ensure product stability and efficacy.
- Support Omics Integration: Correlate lipidomics data with genomics, transcriptomics, and proteomics for comprehensive biological interpretations.
- Evaluate Stress and Disease Markers: Investigate lipid signaling pathways altered under various physiological or experimental conditions, supporting fundamental research and drug development.
Our Service Portfolio: Creative Proteomics' Sphingoid Bases Analysis
- Qualitative Profiling: Identification of known and novel sphingoid bases and derivatives across biological and synthetic samples.
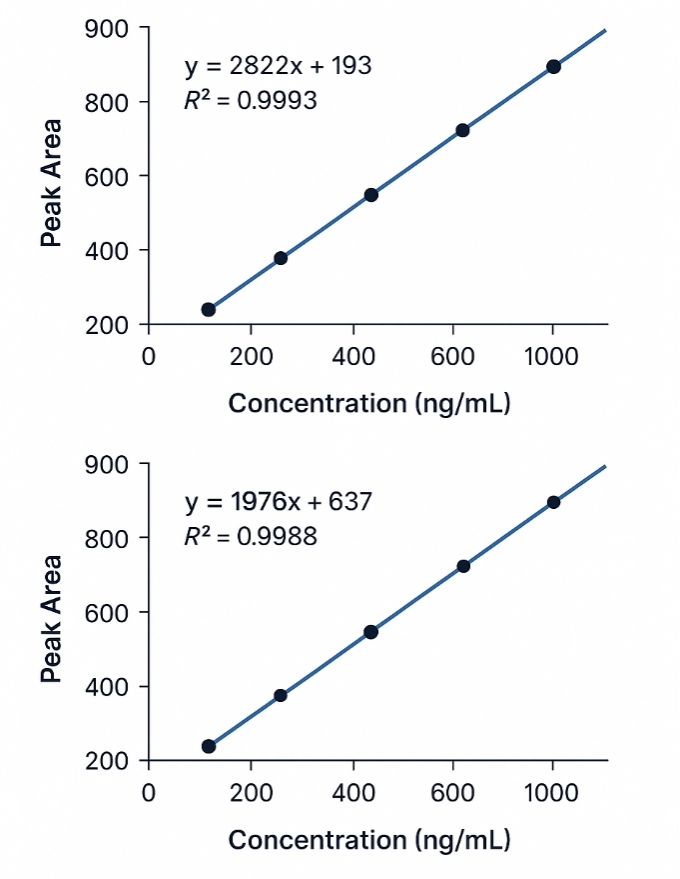
- Absolute Quantification: Precise concentration measurements using internal standards and calibration curves for high accuracy.
- Matrix-Specific Method Development: Custom protocols for serum, plasma, tissues, cell cultures, fermentation broths, plant extracts, and cosmetic products.
- Phosphorylated Sphingoid Bases Detection: Specialized methods for analyzing critical bioactive molecules like sphingosine-1-phosphate (S1P).
- Batch Analysis & High-Throughput Options: Scalable workflows suitable for routine QC, large cohort studies, and high-throughput screening.
- Data Interpretation & Consultation: Comprehensive reports including quantitative data, chromatograms, and expert interpretation for actionable insights.
List of Detected Sphingosine Bases and Related Metabolites
| Analyte Name | Abbreviation | Typical LOD (ng/mL) | Notes |
|---|---|---|---|
| Sphingosine | So | 0.05 | Primary sphingoid base backbone in mammals |
| Dihydrosphingosine (Sphinganine) | dSo | 0.05 | Reduced form of sphingosine |
| Phytosphingosine | PhytoSo | 0.10 | Contains additional hydroxyl group; common in plants |
| Sphingosine-1-phosphate | S1P | 0.10 | Potent signaling lipid |
| Dihydrosphingosine-1-phosphate | dS1P | 0.10 | Phosphorylated derivative of dihydrosphingosine |
| 6-Hydroxy-sphingosine | 6-OH-So | 0.10 | Hydroxylated sphingoid base variant |
| Deoxy-sphingosine | deoxySo | 0.10 | Lacks C1 hydroxyl; linked to certain metabolic disorders |
| Deoxy-dihydrosphingosine | deoxy-dSo | 0.10 | Reduced deoxy variant |
| C20-Sphingosine | C20-So | 0.10 | Longer-chain sphingosine variant |
| C20-Dihydrosphingosine | C20-dSo | 0.10 | Longer-chain reduced variant |
| Sphinganine-1-phosphate | dSo1P | 0.10 | Alternative name for dihydrosphingosine-1-phosphate |
| Sphingadienine | So-diene | 0.10 | Contains two double bonds; rare variant |
| Dihydro-phytosphingosine | dPhytoSo | 0.10 | Reduced phytosphingosine |
| Sphingoid base acetates | Various | 0.10 | Acetylated sphingoid derivatives |
| Sphingoid base glycosides | Various | 0.10 | Glycosylated forms; requires specialized detection |
Advantages of Sphingosine Bases Assay
- Ultra-Low Detection Limits: Achieves limits of detection as low as 0.05 ng/mL for key sphingoid bases, enabling trace-level analysis in complex biological and industrial matrices.
- Exceptional Linearity: Provides calibration curves with R² values typically exceeding 0.999, ensuring accurate quantification across a dynamic range spanning 0.05–1,000 ng/mL.
- High Reproducibility: Delivers coefficient of variation (CV) ≤10% for replicate analyses, supporting consistent results crucial for longitudinal studies or product quality control.
- Matrix Versatility: Validated workflows accommodate diverse samples—including plasma, tissue, fermentation broths, cosmetics, and food—while minimizing matrix effects to less than ±15%.
- Simultaneous Multi-Analyte Detection: Capable of quantifying both free and phosphorylated sphingoid bases within a single LC-MS/MS run, improving efficiency and data comparability.
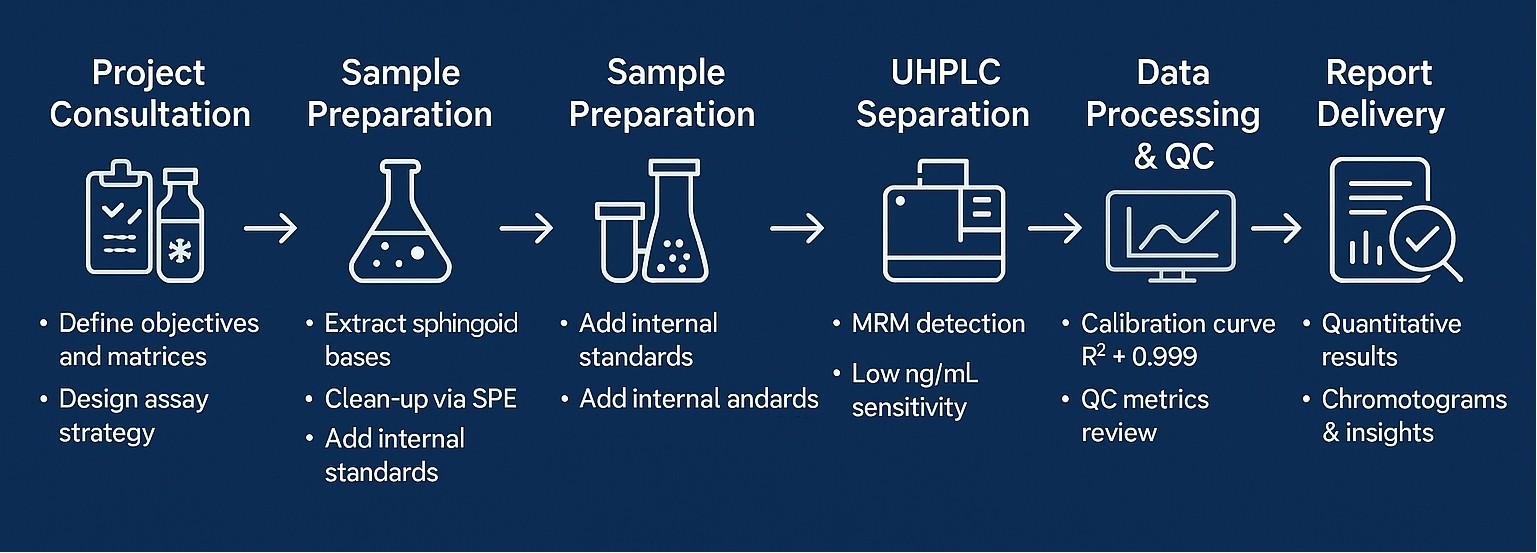
Workflow for Sphingosine Bases Analysis Service
Technology Platform for Sphingosine Bases Analysis Service
LC-MS/MS Platform
- Instrument: Agilent 6495C Triple Quadrupole LC/MS System
- Ionization Mode: Positive Electrospray Ionization (ESI+)
- Operating Mode: Multiple Reaction Monitoring (MRM)
- Dynamic Range: Up to 10⁴ fold (from ~0.05 ng/mL to >1,000 ng/mL)
- Typical LOD: As low as 0.05 ng/mL for key sphingoid bases, depending on matrix and compound
This system offers rapid polarity switching and low noise levels, ideal for differentiating structurally similar sphingoid bases and phosphorylated derivatives.
 Agilent 6495C Triple Quadrupole (Figure from Agilent)
Agilent 6495C Triple Quadrupole (Figure from Agilent)
UHPLC Separation
- UHPLC System: Thermo Scientific™ Vanquish™ or Agilent 1290 Infinity II
- Column: C18, 1.7 µm particle size, 2.1 × 100 mm
- Flow Rate: 0.3 mL/min
- Column Temperature: 40°C
- Mobile Phases: A: Water + 0.1% Formic Acid; B: Acetonitrile or Methanol + 0.1% Formic Acid
- Gradient Program: Optimized to resolve sphingosine, dihydrosphingosine, phytosphingosine, and related analytes with excellent peak shape and retention reproducibility
 Agilent 1260 Infinity II HPLC (Fig from Agilent)
Agilent 1260 Infinity II HPLC (Fig from Agilent)
Key Method Parameters
- Injection Volume: 2–5 µL
- MRM Transitions:
- Sphingosine → product ion at m/z 264.3 → 82.1
- Dihydrosphingosine → m/z 266.3 → 84.1
- Phytosphingosine → m/z 318.3 → 60.1
- Sphingosine-1-phosphate → m/z 380.3 → 264.3
- Additional transitions optimized for rare variants
- Retention Time Stability: <0.2% RSD across batch runs
- Quantification Precision: CV ≤ 10% at low ng/mL levels
Internal Standards
To ensure the highest data quality, we employ isotopically labeled internal standards (e.g., d7-sphingosine, d7-S1P) for:
- Correcting matrix effects
- Enhancing quantification accuracy
Sample Requirements for Sphingosine Bases Analysis Service
| Sample Type | Recommended Volume / Weight | Storage Conditions | Notes |
|---|---|---|---|
| Plasma / Serum | ≥ 200 μL | -80°C | Prefer heparin- or EDTA-treated samples. |
| Cell Culture (Pellet) | ≥ 1 × 10⁶ cells | -80°C | Wash with PBS before freezing. |
| Tissue (Animal / Plant) | ≥ 20 mg | -80°C | Snap-freeze in liquid nitrogen if possible. |
| Fermentation Broth | ≥ 1 mL | -80°C | Filter or centrifuge to remove particulates. |
| Cosmetics / Creams / Oils | ≥ 500 mg | Room temp / Refrigerated | Store away from light and heat. |
| Food / Beverage | ≥ 2 g or ≥ 2 mL | -20°C to -80°C | Provide details on matrix and composition. |
| Standard Solutions | ≥ 100 μL (≥ 1 μg/mL) | -20°C | For custom method validation or calibration. |
Demo Results
FAQ of Sphingosine Bases Analysis Service
Can you analyze phosphorylated sphingoid bases such as sphingosine-1-phosphate (S1P)?
Yes. Our platform includes optimized methods for phosphorylated sphingoid bases like S1P and dihydrosphingosine-1-phosphate, ensuring high sensitivity and selectivity even in complex biological matrices.
Do I need to perform any sample pre-treatment before shipping?
We recommend simple pre-treatment like snap-freezing tissues or centrifuging liquids to remove debris. No further processing is necessary; we perform all extractions and clean-up in-house.
Are internal standards used in your analysis?
Yes. We routinely use isotopically labeled internal standards to correct for matrix effects and enhance quantification accuracy across diverse sample types.
Can your assay distinguish between different sphingoid base chain lengths or structural isomers?
Yes. Our LC-MS/MS methods are optimized for resolving sphingoid bases of varying chain lengths (e.g., C18 vs. C20) and can distinguish cis/trans or saturated/unsaturated variants where applicable.
What format will the analytical report be delivered in?
Our reports include quantitative results, chromatograms, method details, and QC metrics, typically provided in PDF and Excel formats for easy data integration and record-keeping.
Do you accept cosmetic or food product samples for sphingoid bases analysis?
Yes. We frequently analyze sphingoid bases in diverse matrices including cosmetics, creams, oils, and food products, with customized extraction and quantification protocols.
Can you analyze sphingoid bases in lipid extracts prepared by our lab?
Yes. If you have pre-extracted lipid samples, we can analyze them directly. Please provide extraction details, solvents used, and sample concentration to ensure compatibility with our methods.
Is your assay suitable for pharmacokinetic (PK) studies of sphingoid bases?
Yes. Our LC-MS/MS assays offer high sensitivity and reproducibility, making them suitable for PK profiling, including time-course studies, bioavailability assessments, and metabolite quantitation.
Do you offer method transfer or cross-validation if I need to implement the assay in my own lab?
Yes. We can support method transfer by providing detailed protocols, calibration standards, and, if needed, technical training to help your team implement the assay in-house.
Can you quantify both free and phosphorylated sphingoid bases in the same run?
Yes. Our LC-MS/MS methods can simultaneously quantify free sphingoid bases (e.g., sphingosine) and their phosphorylated forms (e.g., S1P) within the same analytical run, optimizing efficiency and consistency.
How small a change in sphingoid base concentration can your assay detect?
Our assay sensitivity allows detection of changes as low as ~10–20% relative differences, depending on the matrix and concentration level. This makes it suitable for studies requiring precise quantitation of subtle metabolic shifts.
Is the assay suitable for large-scale studies or high-throughput screening?
Yes. We support high-throughput analysis, including batch runs for large sample numbers, which is ideal for cohort studies, clinical research, or product batch QC.
Learn about other Q&A about proteomics technology.
Publications
Here are some of lipidomics-related papers published by our clients:

- White matter lipid alterations during aging in the rhesus monkey brain. 2024. https://doi.org/10.1007/s11357-024-01353-3
- Characterization of Dnajc12 knockout mice, a model of hypodopaminergia. 2024. https://doi.org/10.1101/2024.07.06.602343
- Evidence for phosphate-dependent control of symbiont cell division in the model anemone Exaiptasia diaphana. 2024. https://doi.org/10.1128/mbio.01059-24
- The olfactory receptor Olfr78 promotes differentiation of enterochromaffin cells in the mouse colon. 2024. https://doi.org/10.1038/s44319-023-00013-5
- Annexin A2 modulates phospholipid membrane composition upstream of Arp2 to control angiogenic sprout initiation. 2023. https://doi.org/10.1096/fj.202201088R
- Lipid Membrane Engineering for Biotechnology. 2023. https://doi.org/10.48780/publications.aston.ac.uk.00046663