Membrane Proteomics for Accurate Target Discovery and Therapeutic Development
Membrane proteins regulate cell signalling, transport, and communication, making them central to both health and disease. More than 60% of approved drugs act on membrane proteins, yet their hydrophobicity and low abundance make analysis technically demanding.
Creative Proteomics offers a complete membrane proteomics service, covering membrane protein extraction, purification, identification, and structure prediction. Our advanced workflows and curated membrane proteome databases ensure accurate functional insights, supporting target discovery, biomarker research, and therapeutic development.
Key Advantages
- Efficient membrane protein purification with optimised methods
- High-resolution membrane protein identification and structure analysis
- Flexible membrane proteome assay and prediction pipelines
- Access to integrated membrane proteome databases for functional annotation
Submit Your Request Now
×
- Precise membrane protein purification
- Reliable membrane proteome assay
- Advanced membrane protein prediction
- Trusted membrane proteomics expertise
- Overview
- Service & Applications
- Workflow
- Advantages
- Samples & Deliverables
- Case Study
- FAQ & Publications
What Are Membrane Proteins and Why They Matter?
Membrane protein functions are central to life. They regulate the flow of ions and molecules, transmit signals across membranes, and support cell–cell recognition. These proteins are also vital for immune defence, energy conversion, and maintaining cellular structure.
There are several key membrane protein types:
- Integral proteins – span the lipid bilayer and form channels or transporters
- Peripheral proteins – attach loosely to membrane surfaces and mediate interactions
- Lipid-anchored proteins – connect to the membrane via covalently bound lipids
- Chaperone-associated proteins – stabilise or guide folding of other proteins
The membrane proteome represents all membrane proteins within a cell, tissue, or organism. Analysing it helps researchers understand disease mechanisms and cellular communication networks. Because more than half of approved drugs act on membrane proteins, these targets remain at the forefront of therapeutic development.
Abnormal membrane protein structure or expression is strongly associated with cancer, neurodegeneration, and cardiovascular disease. By mapping the membrane proteome array and integrating results with functional assays, researchers can identify biomarkers, validate drug targets, and explore mechanisms of resistance.
Our Membrane Proteomics Service
Creative Proteomics provides a comprehensive membrane proteomics service designed to address the technical challenges of working with hydrophobic and low-abundance proteins. Our platform integrates membrane protein extraction, purification, identification, and bioinformatics-driven prediction, delivering reliable results for research and development.
Core Service Components
- Membrane protein extraction using detergent-based and affinity-based methods optimised for yield and stability.
- Membrane protein purification with advanced chromatography and tag-based enrichment.
- Membrane protein identification through high-resolution LC-MS/MS workflows.
- Membrane proteome assay and array for quantifying expression and comparing samples across conditions.
- Membrane protein prediction and annotation via curated membrane proteome databases and pathway enrichment tools.
Applications of Our Service
- Target discovery – identifying novel therapeutic targets in oncology, neurology, and immunology.
- Biomarker development – detecting disease-associated membrane proteins for translational research.
- Functional biology – exploring protein localisation, structure, and post-translational regulation.
- Resistance mechanism studies – mapping membrane protein differences between drug-sensitive and drug-resistant cells.
For related solutions, see Subcellular Proteomics for receptor analysis, Exosome Proteomics for vesicle-associated targets, and Protein Identification Services for comprehensive characterisation.
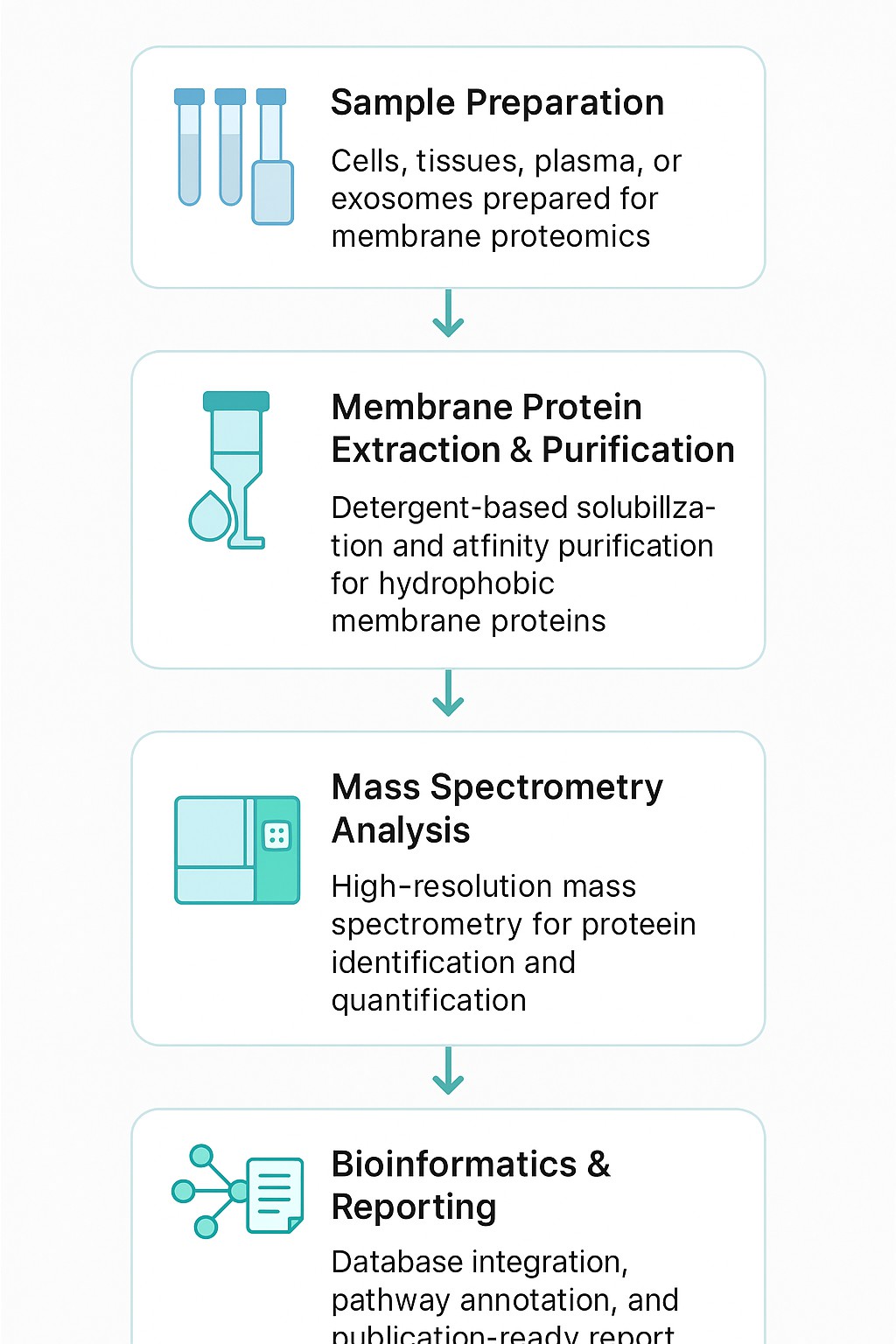
Technical Workflow
Our membrane proteomics workflow has been developed to maximise recovery of hydrophobic and low-abundance proteins while ensuring reproducibility. Each step is supported by validated protocols and advanced instrumentation.
Step 1: Membrane Protein Extraction
We employ detergent solubilisation, affinity capture, and column-based kits to recover membrane proteins while preserving their native conformation. Special care is taken to minimise contamination from cytosolic proteins.
Step 2: Membrane Protein Purification
Affinity purification using tags (His, GST, Rho1D4) or chromatography methods ensures high purity. Optimisation of buffers and detergents improves protein stability, enabling downstream structural or functional analysis.
Step 3: Membrane Proteome Array / Assay
Extracted proteins are analysed using LC-MS/MS with DIA, SWATH, TMT, or iTRAQ quantification. This allows comparative profiling across samples, mapping of membrane protein functions, and integration into a membrane proteome database.
Step 4: Bioinformatics Prediction and Annotation
We provide advanced analysis pipelines covering:
- Subcellular localisation and topology prediction
- Gene Ontology and KEGG pathway enrichment
- Protein–protein interaction network mapping
- Cross-validation with existing membrane proteome arrays and databases
Why Partner with Creative Proteomics
Choosing the right partner is critical for reliable membrane proteomics research. Creative Proteomics combines advanced technology with deep expertise to deliver high-quality, reproducible results.
Comprehensive Expertise
With rich experience in proteomics, our scientists are skilled at handling hydrophobic, low-abundance, and multi-span membrane protein types. We support diverse projects, from academic discovery to pharmaceutical target validation.
High-Quality Deliverables
We provide publication-ready figures, curated bioinformatics reports, and access to curated membrane proteome databases. Each project includes detailed quality control metrics and transparent documentation.
Cutting-Edge Technology
Our platform includes Orbitrap and Q-TOF mass spectrometers, supported by SWATH-MS, DIA, TMT, and iTRAQ quantification. These technologies enable accurate membrane protein identification, structural mapping, and post-translational modification profiling.
Flexible and Customised Solutions
We adapt workflows for different sample types, research questions, and client goals. Options include membrane proteome assays, array-based studies, or predictive modelling for functional insights.
Trusted Global Collaborations
Our services are used by research institutes, CROs, and pharmaceutical companies worldwide. Many clients combine membrane proteomics with Exosome Proteomics, Subcellular Proteomics, Protein-Protein Interaction Analysis or Protein Post-translational Modification Analysis to expand their studies.
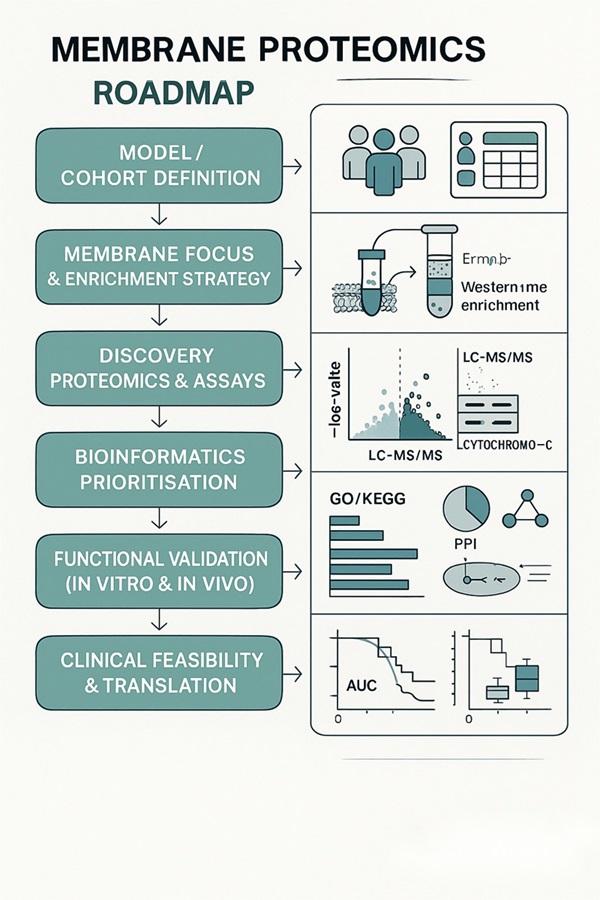
Membrane Proteomics Research Roadmap
Sample Requirements
| Sample Type | Recommended Quantity | Minimum Quantity | Storage & Shipping |
|---|---|---|---|
| Cultured Cells | ≥1 × 10⁷ cells | 5 × 10⁶ cells | Snap-freeze in liquid nitrogen; ship on dry ice |
| Tissue (animal or plant) | 50–200 mg | 20 mg | Remove excess fat/connective tissue; freeze in liquid nitrogen; store at –80°C |
| Plasma/Serum | 500 µL – 10 mL | 200 µL | Centrifuge to remove debris; aliquot and freeze at –80°C |
| Exosomes / Vesicles | ≥1 × 10¹⁰ particles | 5 × 10⁹ particles | Isolate vesicles, resuspend in PBS; freeze at –80°C |
| Bacterial Samples | ≥1 × 10⁹ cells | 5 × 10⁸ cells | Pellet by centrifugation, wash with PBS; freeze at –80°C |
| FFPE Samples | ~80 mg tissue (≥ 4 slides, 5–20 μm) | 40 mg tissue | Transport at room temperature if properly fixed |
General Notes
- Always provide at least one aliquot as backup to avoid freeze–thaw cycles.
- Label tubes clearly (≤4 alphanumeric characters).
- Submit completed electronic and paper submission forms together with QC data.
Deliverables
- Detailed experimental methodology and quality control metrics
- Raw and processed LC-MS/MS data files
- Comprehensive membrane protein identification and quantification tables
- Functional annotations with pathway and network analysis
- Access to curated membrane proteome databases for prediction and comparison
- Publication-ready visual outputs: heatmaps, volcano plots, interaction networks
These deliverables ensure that researchers, CROs, and pharma teams can confidently use results for target discovery, biomarker validation, and mechanistic studies.
Client Case Study: Membrane Proteomics of NK-Cell EVs Defines Cytotoxic Protein Cargo

- Project background
- Client challenges
- Our membrane proteomics solution
- Results
- Conclusions
A client needed a membrane proteomics read-out for extracellular vesicles (EVs) from human NK cells. Their goals were to characterise membrane protein content, verify EV purity, and prioritise candidates for anti-tumour studies.
- Hydrophobic, low-abundance membrane proteins are frequently under-detected.
- EV preparations risk contamination from cytosolic proteins.
- Functional interpretation requires curated membrane proteome annotation.
We applied EV-focused membrane protein extraction, detergent-optimised purification, and high-resolution LC-MS/MS. Downstream analysis used GO/pathway mapping and curated membrane proteome databases to interpret functions and topology.
- EV purity confirmed: Western blot showed enrichment of canonical EV membrane proteins and absence of cytosolic contaminants, validating the extraction workflow.
- Comprehensive proteome profiling: LC-MS/MS identified 623 proteins, including adhesion molecules, signalling receptors, and cytotoxic effectors.
- Functional insights: GO classification revealed EV proteins involved in cell adhesion, immune signalling, and apoptotic regulation, linking membrane cargo to anti-tumour function.
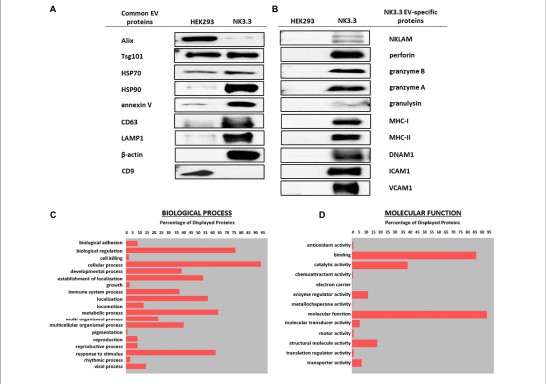 Figure 1. Gene Ontology classification of NK3.3 EV membrane proteome. Biological processes (2C) and molecular functions (2D) highlight adhesion, signalling, and cytotoxic pathways identified via LC-MS/MS.
Figure 1. Gene Ontology classification of NK3.3 EV membrane proteome. Biological processes (2C) and molecular functions (2D) highlight adhesion, signalling, and cytotoxic pathways identified via LC-MS/MS.
By applying our tailored membrane proteomics platform, the client overcame challenges in isolating and profiling EV membrane proteins. The study demonstrated that NK-cell EVs carry a rich repertoire of cytotoxic membrane proteins with therapeutic potential. Our service provided clean datasets, functional interpretation, and a reliable workflow—directly supporting the client's translational research on EV-based cancer therapies.
FAQ
What kinds of membrane proteins can be detected in membrane proteomics analysis?
We can detect integral (transmembrane), peripheral, lipid-anchored, and associated membrane proteins from cells, tissues, exosomes or biofluids; our tools target proteins with one or more hydrophobic transmembrane regions, channels, GPCRs, receptors, and transporters, using extraction + purification to capture both high and low-abundance types.
How do you handle the challenges of low abundance and hydrophobicity in membrane protein extraction?
Our extraction and purification protocols use detergents, affinity tags, organic solvents or special enrichment kits to increase solubility and reduce loss; combined with high-resolution LC-MS/MS techniques and pre-fractionation we overcome suppression and under-representation of hydrophobic proteins in the membrane proteome.
What quantification strategies are available for membrane proteome assays?
We offer both label-based methods (TMT, iTRAQ, SILAC) and label-free options (PRM, SRM, SWATH) to provide comparative quantification; this supports relative expression studies, biomarker discovery, and verification of changes in membrane protein abundance across conditions.
Can you predict membrane protein structure and topology, and what bioinformatics tools are provided?
Yes—our pipeline includes transmembrane region prediction, topology modelling, signal peptide detection, functional annotation, GO/KEGG pathway analysis, interaction network inference, and mapping against curated membrane proteome databases to support predictions of structure and function.
How much sample input and what types of sample are required for reliable membrane proteomics?
We accept a variety of sample types (cultured cells, tissue, plasma/serum, exosomes), and advise sufficient protein quantity and sample purity; poor quality or contaminant-rich samples can reduce coverage and sensitivity.
Is this membrane proteomics service suitable for biomarker discovery and drug target validation?
Yes—our services are designed to discover novel targets, validate membrane proteins as biomarkers, compare disease vs. control states, and feed into follow-up assays or therapeutic development, with robust identification, quantification, and functional insights.
Learn about other Q&A.
Publication

- Extracellular vesicles regulate metastable phenotypes of lymphangioleiomyomatosis cells via shuttling ATP synthesis to pseudopodia and activation of integrin adhesion complexes Kalvala, A. K., Silwal, A., Patel, B., Kasetti, A., Shetty, K., Cho, J. H., ... & Karbowniczek, M. bioRxiv. 2024.
- Tumor-Derived Extracellular Vesicles Regulate the Fate and Function of T-Lymphocyte Via Reprogramming Lipid Metabolism Ma, F. Saint Louis University. 2023.
- Extracellular vesicles from the human natural killer cell line NK3 Cochran, A. M., & Kornbluth, J. Frontiers in Cell and Developmental Biology. 2021.
- Extracellular vesicles secreted by TDO2-augmented fibroblasts regulate pro-inflammatory response in macrophages Peck, K. A., Ciullo, A., Li, L., Li, C., Morris, A., Marbán, E., &am; Ibrahim, A. G. Frontiers in Cell and Developmental Biology. 2021.
Reference
- Rapid Enzyme-Mediated Biotinylation for Cell Surface Proteome Profiling. Anal Chem. 2021 Mar 16;93(10):4542-4551.2. The Cancer Surfaceome Atlas integrates genomic, functional and drug response data to identify actionable targets. Nat Cancer. 2021 Dec;2(12):1406-1422.3. Mass spectrometry and the cellular surfaceome. Mass Spectrom Rev. 2022 Sep;41(5):804-841.












