Phospholipid Analysis Service via Mass Spectrometry
At Creative Proteomics, we offer a highly sensitive and reproducible LC-MS/MS-based phospholipid analysis platform. Designed to meet the needs of both academic and industrial researchers, our solution supports the reliable identification and quantification of phospholipids across various sample types.
Our Phospholipidomics Platform Includes:
- Technology: Advanced Liquid Chromatography–Tandem Mass Spectrometry (LC-MS/MS)
- Approach: Extraction using mixed organic solvents from tissues or cultured cells
- Flexibility: Options for both targeted LC-MS/MS panels and shotgun lipidomics workflows
- Coverage: Comprehensive detection of key phospholipid classes including PC, PE, PS, PI, PG, PA, and cardiolipin
Whether you're mapping metabolic pathways or searching for new disease biomarkers, our phospholipidomics service is built to deliver high-resolution, quantitative data you can trust.
Submit Your Request Now
×- Background
- Methods
- Workflow
- Application
- Metabolite Coverage
- Sample Requirement
- Delivery & Demo
- FAQ
- Case Study
- Publication
What Are Phospholipids?
Phospholipids are a class of complex lipids that contain phosphate groups. They play a central role in forming biological membranes. Structurally, a phospholipid consists of:
- Two long, hydrophobic fatty acid chains (the "tails")
- A hydrophilic phosphate-containing head group
This amphipathic nature allows phospholipids to self-assemble into bilayers in aqueous environments, forming the structural backbone of cell membranes. The phosphate head often contains additional functional groups like amino or alcohol groups, while the tails vary in length and saturation.
Phospholipids are primarily made of carbon (C), hydrogen (H), oxygen (O), nitrogen (N), and phosphorus (P). Key membrane components such as proteins, glycolipids, and cholesterol embed within or attach to this phospholipid bilayer, contributing to membrane fluidity and biological function.
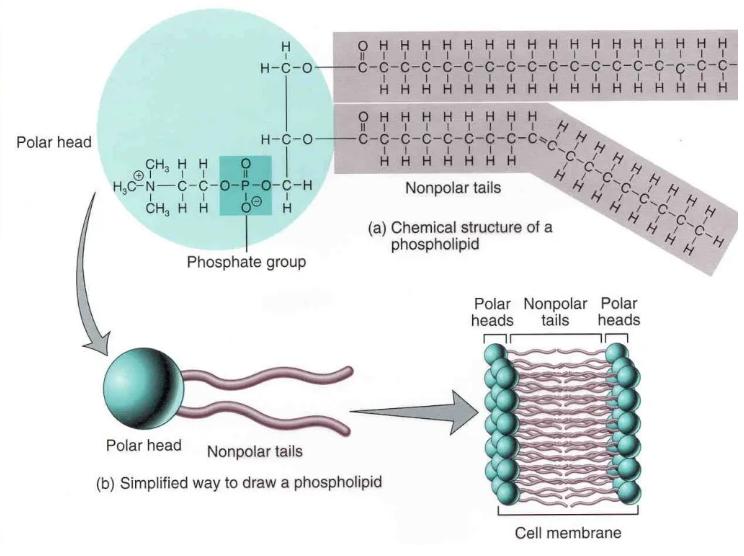 Phospholipid Structure
Phospholipid Structure
Types of Phospholipids
Phospholipids fall into two major categories based on their alcohol backbone:
- Glycerophospholipids – Derived from glycerol
- Sphingolipids (Sphingomyelin) – Based on sphingosine
Glycerophospholipids are the most common in biological systems. Beyond building membranes, they assist in:
- Bile formation
- Lung surfactant composition
- Protein signaling and membrane recognition
Key subclasses include:
- Phosphatidylcholine (PC)
- Phosphatidylethanolamine (PE)
- Phosphatidylserine (PS)
- Phosphatidylinositol (PI)
- Phosphatidylglycerol (PG)
- Phosphatidic acid (PA)
Variants like plasmalogens and lysolecithins arise from different fatty acid chains. One notable type, cardiolipin (CL)—a diphosphatidylglycerol—is found in the mitochondrial inner membrane, where it regulates enzymes involved in oxidative phosphorylation.
Sphingolipids, commonly referred to as sphingomyelins, are especially abundant in neural tissues and play key roles in cell signaling and myelin sheath formation.
Why Phospholipids Matter
Phospholipids do much more than build cell membranes. Disruptions in their metabolism are linked to various conditions, including:
- Metabolic disorders (diabetes, obesity)
- Cardiovascular issues (atherosclerosis, coronary heart disease)
- Neurodegeneration (Alzheimer's disease, brain injury)
- Liver dysfunction and rare syndromes (e.g., Barth syndrome)
- Tumour progression
This makes phospholipid profiling an essential part of biomedical research. By decoding how phospholipids behave in different biological contexts, researchers can:
- Uncover disease mechanisms
- Identify early-stage biomarkers
- Discover novel therapeutic strategies
What Is Phospholipidomics?
First introduced in 2008 by researchers from the University of Bremen, phospholipidomics is a sub-discipline of lipidomics focused on the large-scale analysis of phospholipid species. By mapping phospholipid metabolic networks under various physiological conditions, scientists aim to:
- Identify key phospholipid biomarkers
- Understand how phospholipids regulate metabolism
- Reveal their roles in biological processes
Phospholipidomics relies heavily on Electrospray Ionization-Mass Spectrometry (ESI-MS), which provides high-resolution, high-sensitivity, and high-throughput analysis. Coupling ESI-MS with liquid chromatography (LC-MS) has dramatically improved the field's ability to:
- Detect low-abundance phospholipids
- Differentiate structural isomers
- Discover potential drug targets and lead compounds
This has made phospholipidomics an indispensable tool in biomedical research and pharmaceutical development.
Mass Spectrometry in Phospholipid Analysis
Traditional techniques like HPLC, TLC, and GC-MS offer limited resolution and require time-consuming steps like derivatization. GC-MS, for instance, only provides data on fatty acid chains and lacks full structural insight.
Modern phospholipidomics leverages:
- Shotgun lipidomics – Fast, sensitive, and automated, but struggles with isomer differentiation and ion suppression of low-abundance lipids.
- LC-ESI-MS (Liquid Chromatography-Electrospray Ionization-Mass Spectrometry) – The gold standard for phospholipid analysis, combining effective separation with sensitive detection. This technique excels at high-throughput qualitative and quantitative profiling.
Recent advances in multidimensional mass spectrometry are pushing the boundaries even further, allowing researchers to tackle the complexity and diversity of phospholipid structures with greater precision.
Workflow for Phospholipid LC-MS Analysis Platform
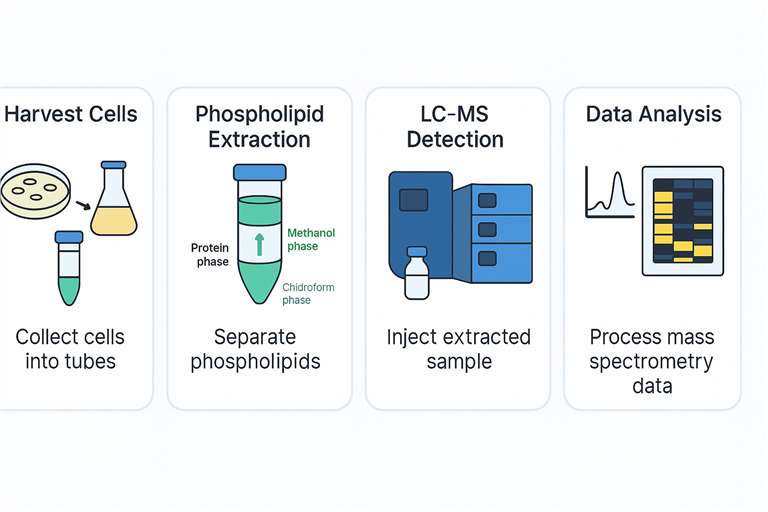
Applications of Phospholipid Mass Spectrometry
As phospholipidomics continues to evolve, mass spectrometry is becoming a powerful tool for uncovering the role of phospholipids in human biology. By precisely measuring shifts in phospholipid composition, this technology is opening new doors across biomedical research and drug development.
Key Applications of Phospholipid LC-MS Analysis:
- Early Detection: Targeted phospholipid profiling helps identify reliable biomarkers. These markers can be linked to disease onset, offering support for intervention strategies.
- Disease Progression Monitoring: Tracking changes in phospholipid levels gives insight into how a disease advances. This enables researchers to assess treatment response or disease dynamics in real time.
- Drug Discovery Support: Understanding how phospholipids shift during disease opens up potential targets for drug design. Mass spectrometry helps map these metabolic changes clearly and accurately.
Comparing phospholipid profiles between healthy and diseased samples also reveals disease-specific patterns. When integrated with enzyme activity data, this analysis helps unravel complex metabolic networks, guiding the development of both diagnostic tools and therapeutic strategies.
As our knowledge deepens, phospholipid mass spectrometry is expected to play a larger role in precision medicine. By tuning phospholipid metabolism, researchers hope to influence disease outcomes in a targeted, systems-level approach.
Phospholipid and Bioactive Lipid Classes Covered
Creative Proteomics' phospholipidomics platform enables precise identification and quantification of a wide range of lipid molecules, including structural lipids, bioactive mediators, and signaling derivatives.
Major Phospholipid Classes
| Lipid Class | Abbreviation |
|---|---|
| Phosphatidic Acid | PA |
| Phosphatidylcholine | PC |
| Phosphatidylethanolamine | PE |
| Phosphatidylserine | PS |
| Phosphatidylinositol | PI |
| Phosphatidylglycerol | PG |
| Cardiolipin | CL |
| Sphingomyelin | SM |
Bioactive Lipid Mediators
| Molecule | Description |
|---|---|
| Platelet-Activating Factor | PAF – inflammatory mediator |
| Lyso-Phospholipids | Lyso-PA, Lyso-PC, Lyso-PS |
| Sphingosine-1-Phosphate | S-1-P – signalling lipid |
Phosphoinositides (PI Derivatives)
| Molecule Name | Abbreviation |
|---|---|
| Monophosphorylated Phosphatidylinositol | PIP |
| Bisphosphorylated Phosphatidylinositol | PIP₂ |
| Trisphosphorylated Phosphatidylinositol | PIP₃ |
Custom Options: We also support profiling of ether-linked phospholipids and other specialised lipid subclasses upon request.
Sample Requirements for Phospholipidomics Analysis
Creative Proteomics supports a wide range of sample types for phospholipid mass spectrometry analysis. To ensure accurate and reproducible results, please prepare your samples according to the guidelines below:
Accepted Sample Types and Minimum Amounts
| Sample Type | Required Amount |
|---|---|
| Animal Tissue | 100–200 mg |
| Plant Tissue | 100–200 mg |
| Plasma/Serum | >100 µL |
| Urine | 200–500 µL |
| Saliva, Amniotic Fluid, Bile, Tears | >200 µL |
| Cultured Cells | >1 × 10⁷ cells |
| Culture Supernatant | >1 mL |
| Wastewater/Culture Medium | >2 mL |
| Microbial Culture | >2 mL |
| Feces/Intestinal Contents | 100–200 mg |
| Soil Samples | >1 g |
| Swabs | 2 swabs |
If your sample type isn't listed above, feel free to contact us for custom evaluation.
Custom Phospholipid Purity Services
Need high-purity phospholipids tailored to your study?
We offer customized phospholipid preparation, involving:
- Rigorous raw material selection
- Targeted extraction and purification
- Fractionation and enrichment
- Precision concentration adjustment to meet your specific research or development goals
This service is ideal for researchers working on lipid-related pathways, bioactive lipid validation, or targeted formulation.
What You'll Receive in the Final Report
We deliver a comprehensive, easy-to-interpret report in both Excel and PDF formats. It includes:
- Raw data and annotated spectra
- Instrument settings and parameters
- Full calculation workflows and quality metrics
Analytes are quantified in µM or µg/mg (for tissue samples), with typical CVs<10%.
Demo
Phospholipid Mass Spectrometry FAQ: What You Need to Know
How do you test for phosphate in lipids?
To detect phosphate groups in lipid molecules, researchers often start by hydrolyzing the sample to release inorganic phosphate. After hydrolysis, the phosphate can be quantified using several analytical methods:
- Colorimetric assays like the ascorbic acid method and Mohr's method rely on the formation of coloured complexes with molybdate, which are measured by UV-Vis spectrophotometry.
- Chromatographic techniques such as HPLC, LC-MS, and ion chromatography enable sensitive separation and quantification.
- NMR spectroscopy offers structural insights, particularly useful for complex lipid mixtures.
- Enzymatic assays may also be used for specific phosphate detection when combined with extraction protocols.
For accurate quantification, samples must be properly extracted—typically using organic solvents—and calibrated against known phosphate standards.
Are phospholipids and amino acids chemically linked?
Yes, in some cases. For instance, phosphatidylserine (PS) includes the amino acid serine as part of its head group. Here, the hydroxyl (-OH) group of serine forms a covalent bond with the phosphate moiety of the phospholipid.
In membrane-associated proteins, hydrophobic amino acids often associate non-covalently with the fatty acid tails of phospholipids. These interactions help anchor proteins into lipid bilayers, even without direct chemical bonding.
What are the main categories of phospholipid MS analysis?
Phospholipid profiling by mass spectrometry (MS) is typically divided into two main approaches:
- Targeted lipidomics: Focuses on predefined lipid species using MRM, SRM, or MS/MS. Ideal for quantifying specific biomarkers, studying cell, or tracing lipid metabolism pathways.
- Untargeted lipidomics: Aims for comprehensive profiling without prior knowledge of the lipid species. Techniques like Shotgun lipidomics or untargeted LC-MS are used to discover new or rare phospholipids in complex samples.
Your choice depends on research objectives—targeted MS is more precise, while untargeted MS is better for broad discovery.
What does phospholipid quantification involve?
Phospholipid quantification is the process of measuring the amount and types of phospholipids in biological samples. This is crucial for:
- Studying metabolic pathways
- Evaluating drug-lipid interactions
- Monitoring cellular responses
Accurate results require careful lipid extraction, proper sample calibration, and validated internal standards. Results are typically reported in micromolar (µM) concentrations or micrograms per milligram of tissue (µg/mg).
How are phospholipids separated in the lab?
Phospholipids are extracted using organic solvents like chloroform and methanol to break down cell membranes and isolate lipids. Separation is then achieved via:
- Liquid chromatography (LC): Excellent for separating lipid classes by polarity.
- Mass spectrometry (MS): Enables precise identification and quantification.
- Shotgun lipidomics: A direct infusion MS approach that provides fast lipid profiling without prior chromatographic separation.
Each method has its strengths depending on the sample type and research goals.
Learn about other Q&A.
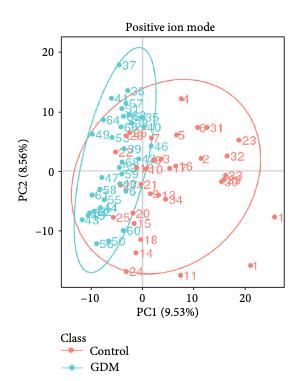
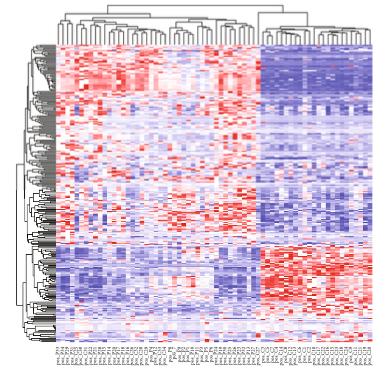
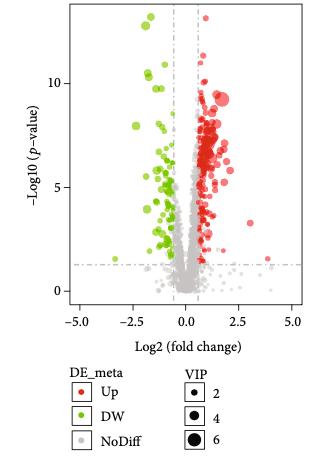
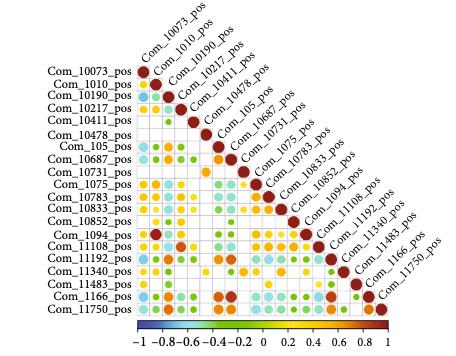
Customer Case Study: LC-MS-Based Phospholipid Profiling in CML Drug Resistance Research

DOI: 10.1002/ctm2.1146
- Background
- How Creative Proteomics Helped
- Results and Impact
- Why It Matters
Chronic myeloid leukemia (CML) remains a clinical challenge due to drug resistance and disease progression despite the success of tyrosine kinase inhibitors (TKIs). In their 2022 study, Gonzalez et al. investigated the tumor suppressor role of the G0/G1 switch gene 2 (G0S2), revealing its impact on lipid homeostasis and therapy resistance. A critical hypothesis in this work was that loss of G0S2 disrupts glycerophospholipid metabolism, thereby contributing to resistance mechanisms in CML.
To test this hypothesis, the research team required a robust, untargeted lipidomics platform capable of sensitive and high-resolution quantification of lipid species, especially glycerophospholipids, in CML cell lines with manipulated G0S2 expression.
Creative Proteomics provided a full-spectrum Phospholipid LC-MS Analysis Service, which included:
- Sample preparation using organic solvent extraction (chloroform:MeOH)
- Lipid class separation with ultra-performance LC (Ultimate 3000 LC, Thermo)
- High-sensitivity detection via electrospray ionization (ESI) and tandem MS
- Comprehensive data processing with LipidSearch™ and multivariate analysis in SIMCA-P
Biological insight through pathway enrichment analysis of significantly altered lipids
The LC-MS analysis identified significant alterations in glycerophospholipid classes—particularly phosphatidylcholine (PC) and phosphatidylethanolamine (PE)—in response to G0S2 expression changes. These changes were critical in linking G0S2 downregulation to disrupted membrane lipid remodeling and therapy resistance in CML.
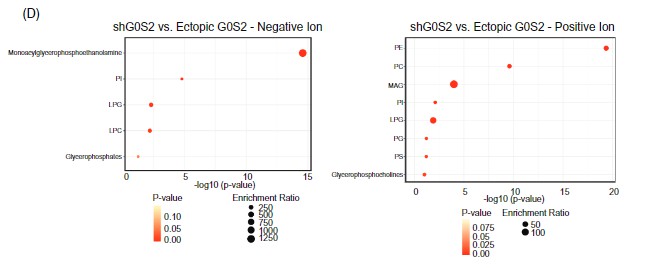 Lipid pathway enrichment analysis reveals glycerophospholipid metabolism as the most dysregulated lipid class upon G0S2 knockdown or overexpression.
Lipid pathway enrichment analysis reveals glycerophospholipid metabolism as the most dysregulated lipid class upon G0S2 knockdown or overexpression.
Key outcome: The lipidomics data provided by Creative Proteomics was pivotal in proving that G0S2 loss drives therapy resistance through dysregulated glycerophospholipid metabolism—a finding with potential therapeutic implications.
This case demonstrates how Creative Proteomics empowers translational cancer research with accurate, publication-grade lipidomics data. By enabling the discovery of a mechanistic link between G0S2, lipid metabolism, and drug resistance, our platform supported the publication of a high-impact study and advanced the understanding of CML pathobiology.
Our Phospholipid Analysis Review

- Timothy M. Sveeggen, Colette A. Abbey, et al,. Annexin A2 modulates phospholipid membrane composition upstream of Arp2 to control angiogenic sprout initiation. The FASEB Journal. 2022. https://doi.org/10.1101/2022.07.07.498997
- Gonzalez, M. A., Olivas, I. M., et al. Loss of G0/G1 switch gene 2 (G0S2) promotes disease progression and drug resistance in chronic myeloid leukaemia (CML) by disrupting glycerophospholipid metabolism. Journal:Clinical and Translational Medicine. 2022. https://doi.org/10.1002/ctm2.1146
- KUMAR, L. K., Han, J., Dalvi, S., Foley, N., Subedi, Y., & Singh, R. Impaired phagocytosis of photoreceptor outer segments by RPE in CLN3 disease is a consequence of altered sphingolipid metabolism. Investigative Ophthalmology & Visual Science. 2024. https://iovs.arvojournals.org/article.aspx?articleid=2796715
Reference
- Sen Yang, Jingyuan Xue, Cunqi Ye.Protocol for rapid and accurate quantification of phospholipids in yeast and mammalian systems using LC-MS.STAR Protocols. 2022