Sphingosine-1-phosphate (S1P) Quantification Service
Accurate analysis of sphingosine-1-phosphate (S1P) is critical for lipidomics, immunology, inflammation, and vascular biology. Creative Proteomics offers a robust, high-sensitivity LC-MS/MS platform that enables reliable S1P quantification, even in ultra-low abundance samples.
Our advanced analytical workflow helps you:
- Precisely quantify S1P in plasma, tissues, cells, and CSF
- Differentiate S1P from structurally similar sphingolipids
- Support biomarker discovery, metabolic flux studies, and pathway profiling
- Ensure reproducibility and batch stability across your experiments
Explore our capabilities below and see what sets us apart.
Submit Your Request Now
×
What You Receive
- Quantitative reports with absolute concentrations (nM/pM)
- Chromatograms and MRM transitions
- Batch-to-batch reproducibility data
- Interpretation-ready data tables (Excel/PDF)
- Optional: pathway mapping visualizations
- What We Provide
- Advantages
- Technology Platform
- Sample Requirement
- Demo
- FAQs
What is Sphingosine-1-phosphate (S1P)?
Sphingosine-1-phosphate (S1P) is a potent bioactive lipid metabolite derived from sphingolipid metabolism. Functioning as a crucial signaling molecule, S1P regulates a wide range of biological processes, including cellular proliferation, migration, survival, vascular integrity, and immune cell trafficking. Due to its involvement in lipid signaling networks, S1P has become a key target in studies related to inflammation, metabolic regulation, neurodegenerative disorders, and immunological responses.
However, its amphipathic nature, low abundance, and tight biological regulation demand robust, high-resolution analytical technologies for accurate quantification and profiling.
Why Choose Our S1P Analysis Service?
Sphingosine-1-phosphate analysis is often challenged by its complex biological matrices, structural similarities with related sphingolipids, and susceptibility to degradation. Our service is designed specifically to address these challenges.
Creative Proteomics offers a targeted, quantitative, and highly sensitive S1P analysis platform that enables researchers to:
- Precisely quantify S1P in a variety of biological samples including plasma, serum, tissues, and cultured cells.
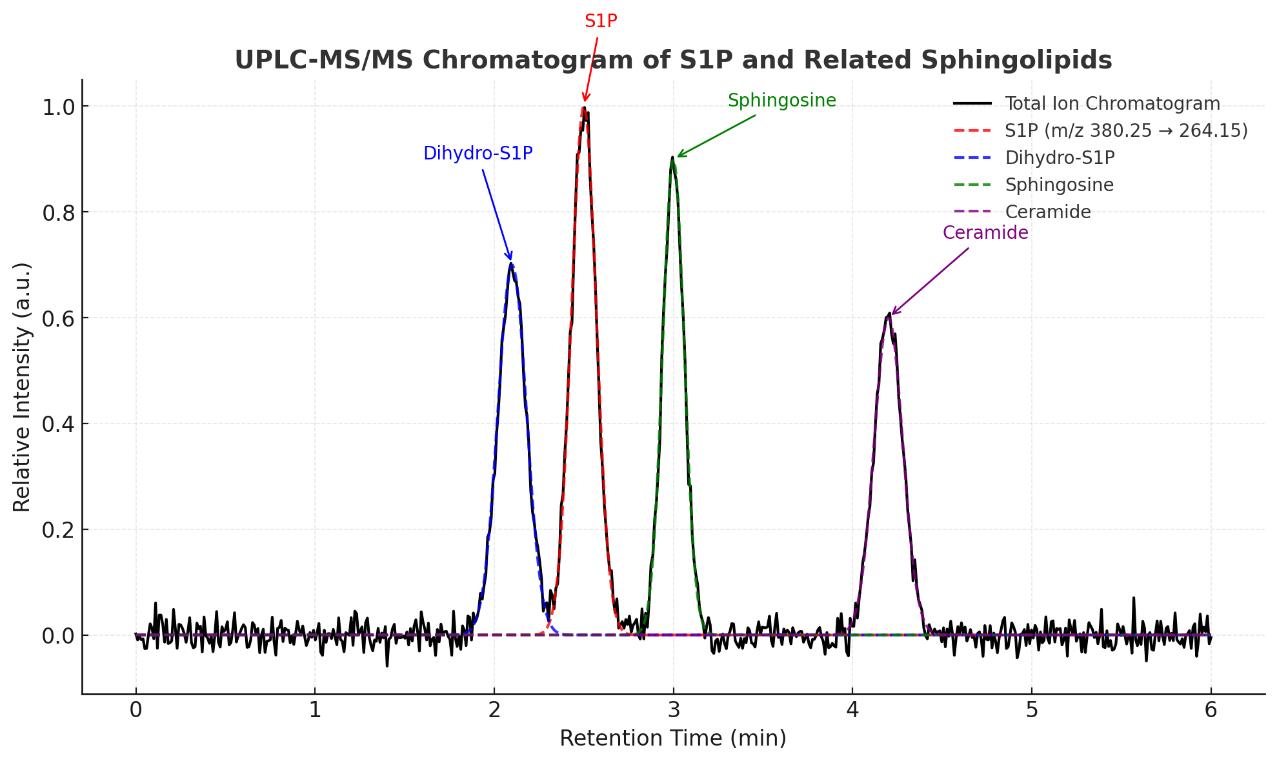
- Identify and differentiate structurally similar sphingolipids such as sphingosine, ceramides, and sphinganine-1-phosphate.
- Support lipidomic profiling in pharmacological and biomarker studies, helping to uncover dynamic lipid signaling pathways.
- Ensure analytical reproducibility across batches, enabling longitudinal and comparative studies with confidence.
What Specific Services Are Included in Our S1P Analysis Package
High-Resolution Quantification of S1P Across Multiple Sample Types
Optimized for low-volume, low-abundance samples such as cerebrospinal fluid (CSF), tumor lysates, immune cells, and exosomes. Supports ultra-trace quantification in difficult matrices.
Time-Course and Treatment-Response S1P Profiling
Enables kinetic studies to monitor S1P dynamics under drug exposure, immune activation, or cellular stress, with batch-stable analytical repeatability.
Differential Sphingolipid Profiling Across Biological Conditions
Comparative profiling of S1P and related sphingolipids across disease states, treatment groups, or tissue types to support biomarker screening or mechanistic research.
S1P Pathway Mapping with Targeted Panel Customization
Custom-configured panels to investigate S1P metabolic flux, including enzymes or intermediates such as SphK1/2 substrates, ceramides (C16–C24), and sphingomyelins.
Isomer-Specific Detection of Sphingolipid Subspecies
Advanced separation and quantification of structurally similar S1P analogs or ceramide isomers, supporting structural-functional studies.
Species-Specific S1P Profiling (Human, Mouse, Rat, etc.)
Validated protocols for commonly used model organisms, ensuring biological relevance and translational accuracy.
Support for Immunological and Vascular Research Models
Tailored to studies on T-cell trafficking, endothelial permeability, and inflammatory signaling, where S1P plays a regulatory role.
List of Detected Sphingosine-1-phosphat and Related Metabolites
| Analyte Name | Classification | Notes / Relevance |
|---|---|---|
| Sphingosine-1-phosphate (S1P) | Bioactive sphingolipid | Key signaling molecule in immune and vascular systems |
| Dihydro-Sphingosine-1-phosphate | S1P analog | Saturated form, involved in similar signaling |
| Sphingosine | Precursor | Direct precursor to S1P via sphingosine kinase |
| Dihydrosphingosine (Sphinganine) | Precursor | Precursor in de novo sphingolipid synthesis |
| Ceramide (C14:0 – C26:1) | Metabolic intermediate | Downstream of sphingosine; includes diverse fatty acyl chains |
| Ceramide-1-phosphate | Functional analog | Pro-inflammatory sphingolipid; overlaps with S1P function |
| Sphingomyelin (various chain lengths) | Structural sphingolipid | Major membrane component; source of ceramide |
| Lactosylceramide | Glycosphingolipid | Implicated in inflammation and cancer signaling |
| Glucosylceramide | Glycosphingolipid | Important in Gaucher disease and other lysosomal disorders |
| Galactosylceramide | Glycosphingolipid | Relevant in neural tissue; demyelination research |
| Hexosylceramide (C16:0 – C24:1) | Glycosphingolipid | Includes GlcCer and GalCer; used in metabolic profiling |
| Phytosphingosine | Sphingoid base | Plant- and yeast-derived analog; sometimes endogenous |
| Dihydroceramide | Biosynthetic intermediate | Precursor of ceramide; involved in ER stress responses |
| 3-Ketosphinganine | Early precursor | Product of serine palmitoyltransferase (SPT) |
| Sphingosine phosphate lyase substrate | Pathway intermediate | Reflects S1P degradation flux |
| Sphingosine kinase 1/2 substrates | Enzymatic substrates | For SphK1/2 activity mapping |
| Lysosphingomyelin | Lysosphingolipid | Catabolic product; accumulates in lysosomal storage disorders |
| Lysophosphatidic acid (LPA) | Functional analog (non-sphingolipid) | Cross-talks with S1P receptors |
| C1P/Ceramide ratio | Derived metric | Used to evaluate inflammatory vs. apoptotic sphingolipid signaling |
| S1P/Ceramide ratio | Derived metric | Marker for cell fate decisions |
Advantages of Sphingosine-1-phosphat Assay
- Ultra-High Sensitivity: Detects S1P at sub-picomolar levels (<0.5 pM), enabling accurate measurement of low-abundance lipids.
- Wide Dynamic Range: Quantifies S1P across 0.1 nM to 1000 nM, accommodating diverse sample concentrations without extra processing.
- High Precision: Maintains intra- and inter-assay CVs below 8%, ensuring consistent results across batches.
- High Throughput: Processes over 100 samples daily with rapid turnaround.
- Comprehensive Profiling: Simultaneously measures 20+ sphingolipid species for detailed pathway analysis.
- Matrix Compatibility: Validated for plasma, serum, CSF, tissues, and cells with recovery rates >85%.
- Accurate Quantification: Internal standards and calibration curves yield data within ±10% accuracy.
Workflow for Sphingosine-1-phosphat Analysis Service
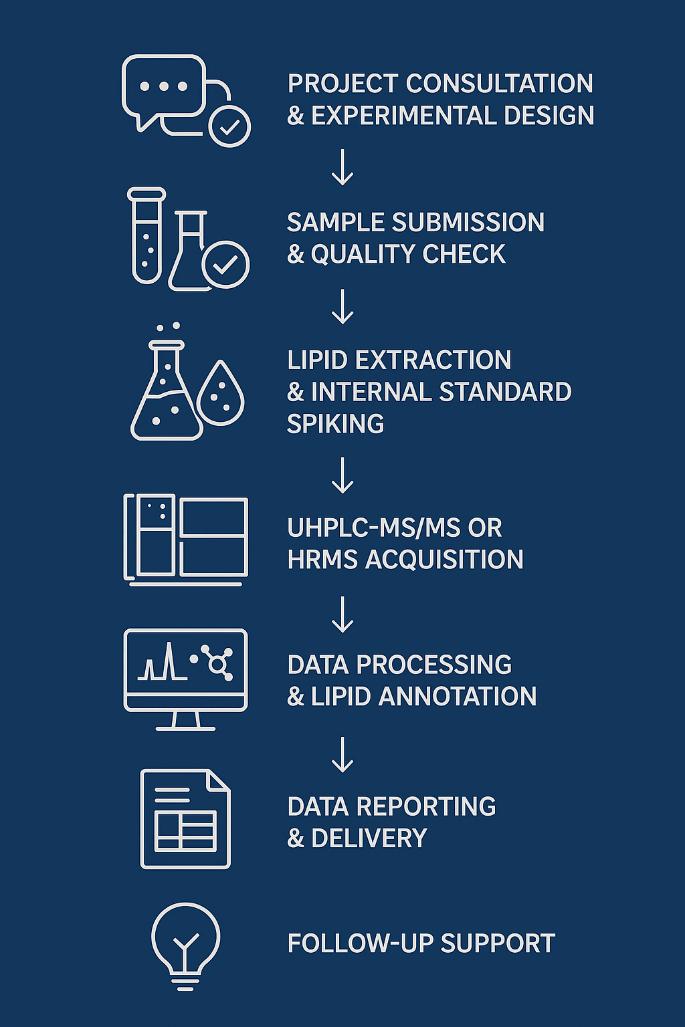
1. Project Setup
Define sample type, analytical goals, and target sphingolipids. Configure internal standards and calibration range to match expected concentration levels.
2. Sample Check-In & Pre-QC
All samples are verified for labeling, volume, and integrity. Logged into LIMS with traceable IDs and processed under controlled thawing conditions.
3. Lipid Extraction
Matrix-specific protocols are applied for deproteinization and organic extraction, with antioxidants and isotope-labeled S1P standards added to ensure quantification accuracy.
4. UPLC-MS/MS Analysis
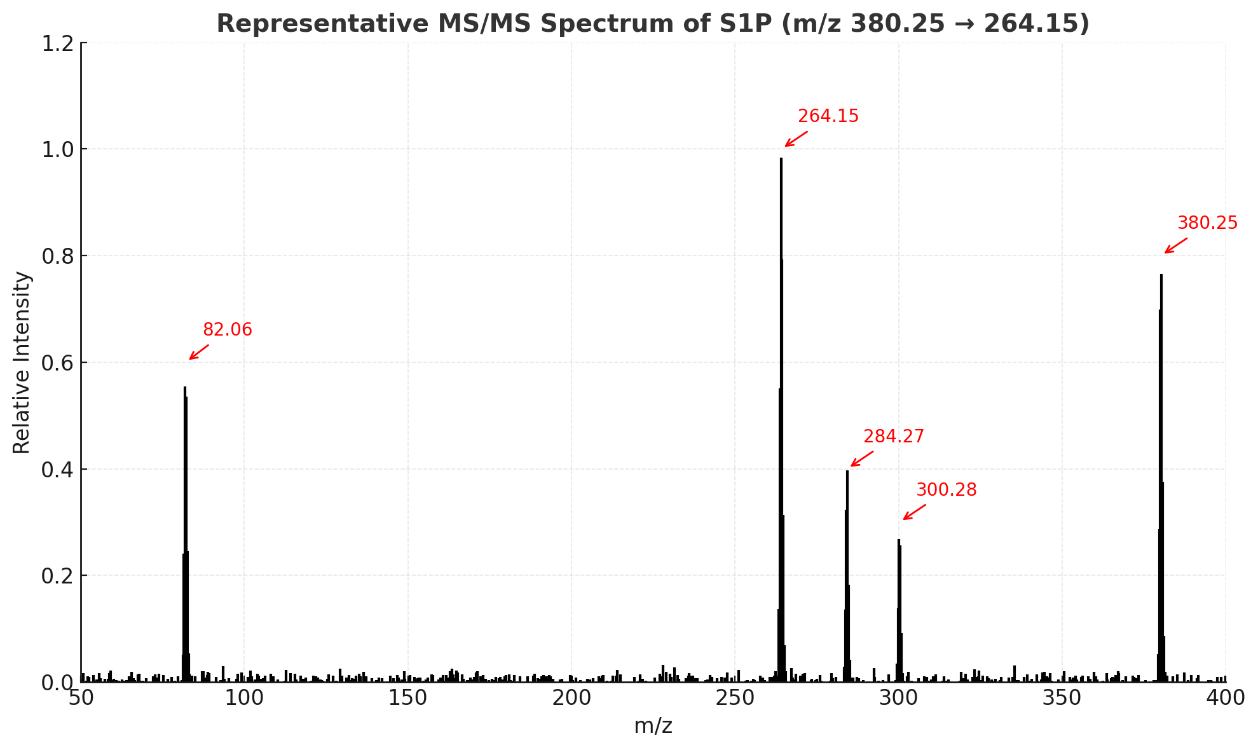
High-resolution chromatographic separation and MRM-based mass spectrometry enable sensitive detection and isomer resolution of S1P and related lipids.
5. Data Processing & Reporting
Quantification is performed using validated calibration curves. Reports include raw and normalized data, QC metrics, and pathway-relevant lipid ratios (e.g., S1P/ceramide).
 Vanquish™ UHPLC (Figure from Thermo Scientific)
Vanquish™ UHPLC (Figure from Thermo Scientific)
What Analytical Methods and Instruments Do We Use?
Ultra-Performance Liquid Chromatography (UPLC):
- Instrument: Waters ACQUITY UPLC I-Class System
- Column: BEH C18, 2.1 × 100 mm, 1.7 µm particle size
- Mobile Phase: Gradient elution using water and acetonitrile with 0.1% formic acid
- Flow Rate: Typically 0.3 mL/min
- Injection Volume: 2–5 µL
 Waters ACQUITY UPLC System (Figure from Waters)
Waters ACQUITY UPLC System (Figure from Waters)
Tandem Mass Spectrometry (MS/MS):
- Instrument: AB Sciex Triple Quad™ 6500+ or Thermo TSQ Altis™
- Ionization Mode: Positive Electrospray Ionization (ESI+)
- Acquisition Mode: Multiple Reaction Monitoring (MRM) for targeted detection
- Key Parameters:
- Curtain Gas: ~30 psi
- Ion Spray Voltage: ~5500 V
- Temperature: 400–500 °C (source dependent)
- Collision Energy: Optimized per analyte (typically 25–40 eV)
- Dwell Time: 20–50 ms per MRM transition
 SCIEX Triple Quad™ 6500+ (Figure from Sciex)
SCIEX Triple Quad™ 6500+ (Figure from Sciex)
Analytical Performance
- Limit of Detection (LOD): Sub-picomolar (low femtomole range) sensitivity for S1P in plasma and tissue samples.
- Linearity: Quantification over a broad dynamic range (typically 0.1 to 1000 nM).
- Precision: Intra- and inter-day coefficient of variation (CV) <10%.
- Throughput: Capable of analyzing >100 samples per day with robust reproducibility.
Sample Requirements for Sphingosine-1-phosphate Analysis Service
| Sample Type | Minimum Volume | Storage Conditions | Notes |
|---|---|---|---|
| Plasma | ≥ 100 µL | Store at -80°C; EDTA or heparin anticoagulant preferred | Avoid repeated freeze-thaw cycles |
| Serum | ≥ 100 µL | Store at -80°C | Clotting time: 30 min at room temp |
| Tissue (fresh/frozen) | ≥ 20 mg | Store at -80°C, snap-frozen in liquid nitrogen | Homogenize before extraction |
| Cell Pellet | ≥ 1 × 10^6 cells | Store at -80°C | Wash cells with PBS before freezing |
| Cerebrospinal Fluid (CSF) | ≥ 200 µL | Store at -80°C | Use polypropylene tubes |
| Other Biofluids | ≥ 200 µL | Store at -80°C | Contact for specific requirements |
Demo Results
FAQ of Sphingosine-1-phosphate Analysis Service
Can this assay differentiate IAA from structurally similar auxins or precursors?
Yes. Our platform utilizes targeted LC-MS/MS with optimized chromatographic separation and MRM transitions to distinguish IAA from closely related molecules such as indole-3-butyric acid (IBA), indole-3-pyruvic acid (IPyA), and tryptophan-derived precursors.
Is the method suitable for both plant and microbial samples?
Absolutely. Our assay is validated across a wide range of biological matrices, including plant tissues (roots, shoots, leaves), culture supernatants, and microbial fermentation samples, making it suitable for studies involving rhizosphere signaling or endophytic bacteria.
How is sample integrity maintained for accurate IAA measurement?
We use acidified extraction buffers and rapid sample processing to minimize enzymatic degradation and oxidation of IAA. Additionally, internal standards are added early in the workflow to correct for sample loss and matrix effects.
Can you support dose-response or hormone treatment studies?
Yes. We can accommodate experimental designs involving hormone treatments, auxin antagonists, or transgenic lines, with precise quantification across multiple time points or conditions for comparative analysis.
Is it possible to co-detect other plant hormones in the same assay?
Upon request, we offer multiplexed hormone profiling services. IAA can be quantified alongside other phytohormones such as abscisic acid (ABA), gibberellins (GA), salicylic acid (SA), and jasmonic acid (JA), depending on the study design and detection requirements.
What quality controls are in place to ensure result reliability?
Each batch includes system suitability checks, matrix-matched calibration curves, internal standards, and pooled quality control (QC) samples to ensure analytical reproducibility and quantitative accuracy.
Can this assay be applied to low-biomass or rare tissue types?
Yes. Our extraction and detection protocols are optimized for high sensitivity, allowing accurate IAA quantification from as little as 5–10 mg of plant tissue or <100 µL of liquid samples, including meristems or micropropagated materials.
Do you offer consultation for experiment setup or data interpretation?
We provide pre-assay consultation to optimize sampling and experimental design, as well as post-assay technical support to help interpret hormone flux, metabolic shifts, or pathway insights based on your IAA data.
Learn about other Q&A about proteomics technology.
Publications
Here are some of lipidomics-related papers published by our clients:

- White matter lipid alterations during aging in the rhesus monkey brain. 2024.
- Characterization of Dnajc12 knockout mice, a model of hypodopaminergia. 2024.
- Evidence for phosphate-dependent control of symbiont cell division in the model anemone Exaiptasia diaphana. 2024.
- The olfactory receptor Olfr78 promotes differentiation of enterochromaffin cells in the mouse colon. 2024.
- Annexin A2 modulates phospholipid membrane composition upstream of Arp2 to control angiogenic sprout initiation. 2023.
- Lipid Membrane Engineering for Biotechnology. 2023.