Accurate S-Nitrosylation Site Mapping Service Using LC-MS/MS and TMT Labeling
At Creative Proteomics, we combine advanced instrumentation with deep scientific expertise to deliver accurate, reproducible, and high-throughput S-nitrosylation site mapping.
- Advanced Instrumentation: We leverage the Thermo Fisher Easy-nLC 1000 and LTQ Orbitrap ETD for high-resolution S-nitrosylation analysis.
- Optimized Workflow: Our refined protocol ensures faster turnaround and increased sensitivity in mapping nitrosylation sites.
- End-to-End Support: From sample prep to data analysis, we manage every step.
Submit Your Request Now
×- Background
- Methods
- Workflow
- Advantage
- Platform
- Sample Requirement
- Demo
- FAQ
- Case Study
- Publication
What is S-Nitrosylation?
S-nitrosylation is a reversible post-translational modification of a cysteine residue, which is mediated by nitric oxide (NO). To date, more than 3000 proteins have been identified in cell as being S-nitrosylated. As an important post-translational modification, S-nitrosylation modification can change the conformation, activity and interaction of proteins, and thus regulate cell signalling, redox balance, immune response, vascular function and cell cycle. In addition, it has an important role in physiological processes such as vasodilation, nerve conduction, cell growth and division, and also closely affects the occurrence, development and progression of cardiovascular diseases, neurodegenerative diseases, cancer and other diseases.
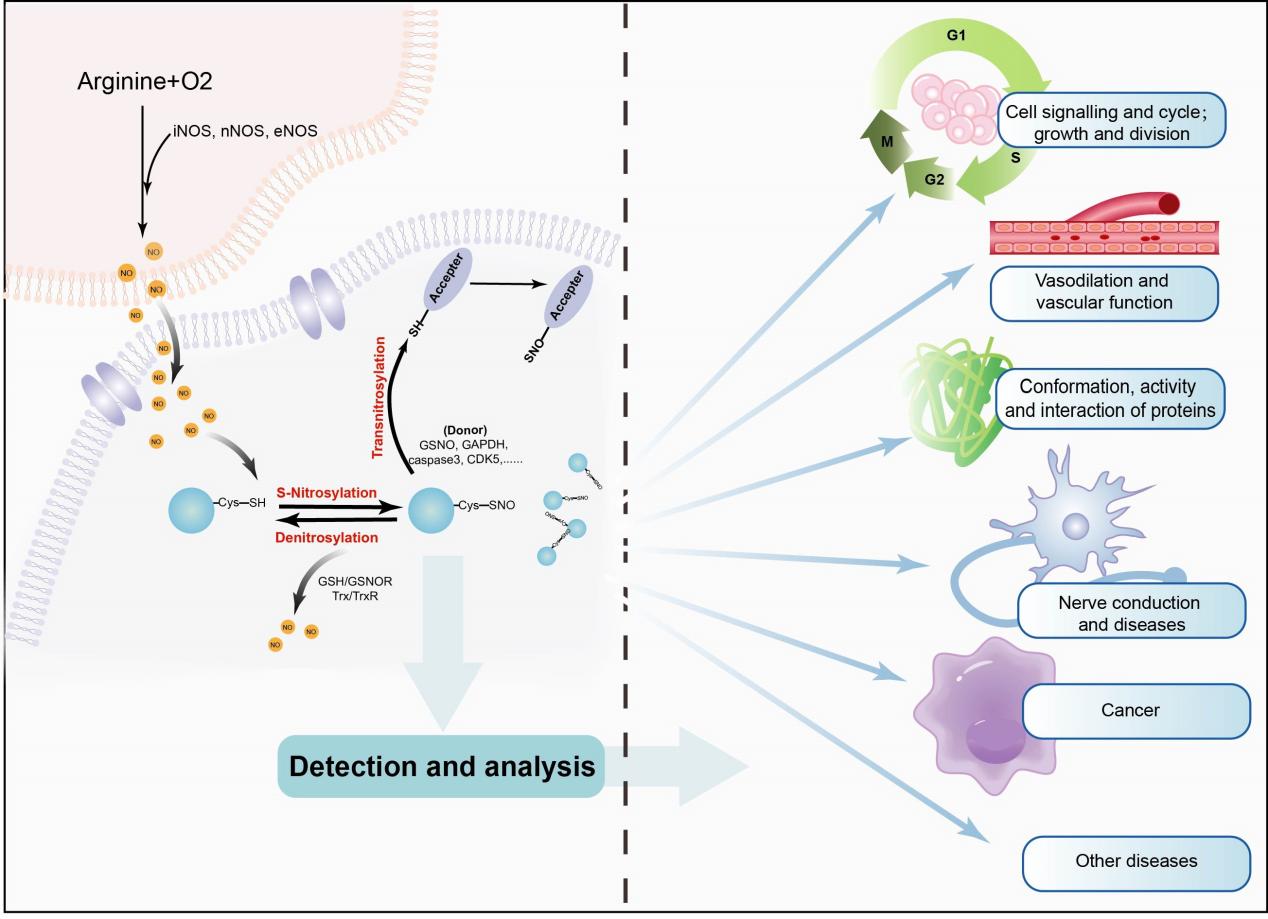 Figure 1:The occurrence and effect of S-Nitrosylation.
Figure 1:The occurrence and effect of S-Nitrosylation.
Mechanism of S-Nitrosylation
S-nitrosylation is a naturally occurring modification where NO attaches to specific cysteine residues in proteins. This reaction is driven by NO generated by nitric oxide synthases (NOS) within mammalian cells. Depending on the number of cysteines involved, S-nitrosylation can occur at a single site (mono-S-nitrosylation) or at multiple sites (poly-S-nitrosylation).
Not all cysteines are equally susceptible. For a cysteine to be S-nitrosylated, it must meet several conditions:
- Be located near NOS
- Reside within a specific acidic or basic motif
- Be found in a hydrophobic region of the protein
- Be physically accessible (i.e., not buried or blocked)
From Local Events to Systemic Effects: Transnitrosylation
Once the initial S-nitrosylation occurs, the NO signal can be passed on to other proteins through transnitrosylation—a process that spreads the modification beyond the original target. This transfer relies on charged amino acid motifs that promote strong electrostatic interactions between the donor (S-nitrosylated protein) and the acceptor protein.
Key players in this cascade include known S-nitrosylases, which donate the NO group:
- S-nitrosoglutathione (GSNO)
- Haemoglobin
- Thioredoxin
- GAPDH
- Caspase-3
- CDK5
These donor proteins have a higher redox potential, allowing them to transfer the NO group and become denitrosylated in the process.
Keeping the Balance: Denitrosylation
The modification is reversible. Denitrosylation removes NO groups from proteins through enzyme-catalysed reactions, maintaining cellular redox homeostasis. Two well-characterized enzymes carry out this function:
- Thioredoxin/thioredoxin reductase (Trx/TrxR)
- Glutathione/S-nitrosoglutathione reductase (GSH/GSNOR)
Together, S-nitrosylation and denitrosylation form a regulatory cycle. Their interplay determines the overall level of protein S-nitrosylation and, consequently, affects various signalling pathways across the cell.
Our S-Nitrosylation Site Mapping Service
S-nitrosylation can dramatically alter protein behaviour, from modulating enzymatic activity to rewiring entire signalling pathways. However, to truly understand its biological role, it's critical to pinpoint exactly where these modifications occur—and what they do. That's where Creative Proteomics comes in.
We've developed a high-sensitivity LC-MS/MS-based workflow designed to identify and characterize S-nitrosylated cysteines in both eukaryotic and prokaryotic samples. Our platform enables researchers to uncover detailed, site-specific insights into this dynamic post-translational modification.
Advancing S-Nitrosylation Detection: From Biotin-Switch to TMT Labeling
Understanding NO signaling requires accurate identification of S-nitrosylated cysteine (SNO-Cys) sites. Over the years, various detection methods have been developed, with the Biotin Switch Technique (BST)—first introduced by Jaffrey in 2001—being one of the most widely used.
BST works by chemically converting SNO modifications into biotin-labeled sites, which are then detected using biotin-specific antibodies. While effective, this method presents several challenges:
- Low throughput, making it unsuitable for large-scale studies
- Limited site resolution, complicating precise localization of modified cysteines
- Risk of false positives, due to interference from endogenous biotin-like molecules during affinity enrichment
To overcome these limitations, we've adopted an advanced TMT labeling strategy. Using 6-plex Tandem Mass Tags (TMT), our approach allows for:
- Simultaneous identification and quantification of S-nitrosylation events
- Improved detection specificity and accuracy
- Scalable, multiplexed analysis across multiple biological samples
This upgraded method enhances both the depth and reliability of SNO-Cys site mapping—critical for uncovering the true complexity of NO-mediated signaling.
How the TMT Labeling Strategy Works
The TMT reagent offers a highly selective way to label cysteine-containing peptides from complex biological samples. It binds irreversibly to free thiol groups on cysteine residues, allowing researchers to confidently map redox-related modifications like S-nitrosylation.
To enrich for these labeled peptides, we use a two-step approach:
- Affinity enrichment using immobilized anti-TMT antibody resin
- Targeted elution with a dedicated TMT elution buffer
This strategy mirrors the concept behind ICAT™ (Isotope-Coded Affinity Tag) techniques, combining selective thiol tagging with high-affinity capture to streamline the workflow.
What Makes TMT Unique?
Each reagent in the iodoTMT-plex kit shares the same parent mass but differs by:
- A Cysteine-Reactive Group for thiol-specific binding
- A Mass Normalizer to ensure accurate quantification
- A Mass Reporter for multiplexed sample analysis
With the ability to label up to six different biological samples in a single run, TMT reagents significantly reduce sample complexity, expand the detectable dynamic range, and provide a robust platform for S-nitrosylation research.
Workflow of Our S-Nitrosylation Analysis Service
Classically, the workflow of our S-nitrosylation includes protein extraction, Biotin-Switch Assay and enrichment of S-nitrosylated peptides, identification of S-nitrosylation sites, and relative quantification of changes for identified S-nitrosylation sites. Additionally, the application of MS analysis can be complemented by incorporating TMT labeling techniques to enhance the precision of relative quantitation between samples.
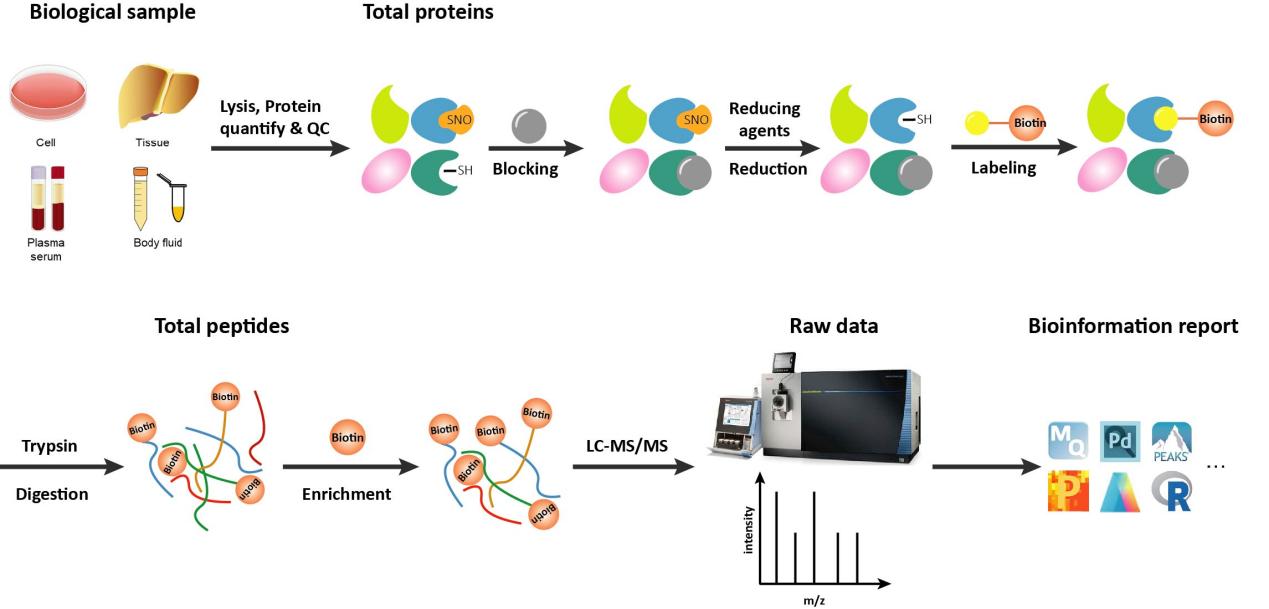
 Figure 2: Workflow of our S-Nitrosylation analysis service.
Figure 2: Workflow of our S-Nitrosylation analysis service.
- Sample lysis and protein extraction.
- All free sulfhydryl group are blocked.
- The S-nitrosylated cysteine are reduced by ascorbic acid.
- The free sulfhydryl group are switched by biotin.
- Protein digestion and biotinized peptide enrichment
- Biotinized peptide were detected by LC-MS/MS.
- Raw data interpretation and bioinformatics analysis
Technological Advantages That Set Us Apart
Expert-Led, Data-Driven
Our team of seasoned PTMs specialists ensures every project benefits from:
- Rigorous quality control protocols
- Professional data pre-processing and interpretation
- Ultra-high-resolution mass spectrometry for precise detection
Consistency You Can Trust
We guarantee high intra- and inter-assay reproducibility, providing the confidence you need in downstream data analysis and decision-making.
Multiplex, High-Throughput Capability
Our optimized workflow delivers comprehensive coverage of S-nitrosylation sites—ideal for large-scale proteomics projects or pathway-level investigations.
Cutting-Edge Instrumentation
We utilize industry-leading platforms to enhance sensitivity and resolution, including:

Exactive

Orbitrap Fusion™ Tribrid™
With these advantages, Creative Proteomics offers unmatched precision and depth for your S-nitrosylation studies.
Samples Requirement for S-Nitrosylation Analysis
| Sample | Sample Quantity | |
|---|---|---|
| Tissue | animal tissue | 200-500mg |
| fresh plant | 200-500mg | |
| Cell | suspension cell | > 3x107 |
| adherent cell | > 3x107 | |
| microorganism | > 50 mg or 3 × 107 cells | |
| Body fluid | Serum/plasma | 1mL |
| Protein | Total protein > 2mg and concentration >1 μg/μL | |
Note: In order to ensure the test results, please inform the buffer components if you give us proteins, whether it contains thiourea, SDS, or strong ion salts. In addition, the sample should not contain components such as nucleic acids, lipids, and polysaccharides, which will affect the separation effect.
Results Delivery & Demo
- Detailed report, including experiment procedures, parameters, etc.
- Raw data and data analysis results.

FAQ: S-Nitrosylation Site Mapping Service
What sample types can be used for S-nitrosylation site analysis?
We accept a wide range of biological samples, including animal tissues, plant tissues, cultured cells (adherent or suspension), microorganisms, serum/plasma, and total protein extracts. Please refer to our sample requirement table to ensure optimal results.
What is the minimum amount of protein required for analysis?
We require at least 2 mg of total protein with a concentration of ≥1 µg/μL. Please inform us of any buffer components that may interfere with the labeling process (e.g., SDS, thiourea, high-salt).
How do TMT labeling and Biotin Switch techniques differ? Which one should I choose?
The Biotin Switch method is a classic approach for detecting SNO sites, while TMT labeling offers higher throughput, quantitative accuracy, and multiplexing capacity (up to 6 samples in parallel). We can recommend the best approach based on your research goals and sample availability.
Can you analyze low-abundance S-nitrosylation sites?
Yes. Our workflow incorporates affinity enrichment and ultra-high-resolution LC-MS/MS, enabling detection of low-abundance, site-specific SNO modifications with high sensitivity.
How long does the entire analysis take?
Turnaround time depends on the sample type, project complexity, and chosen detection method. We prioritize efficiency without compromising data quality, and expedited services are available upon request.
What kind of data and results will I receive?
You will receive a comprehensive report including:
- Experimental workflow and parameters
- Raw LC-MS/MS data files
- Identified S-nitrosylated peptides and corresponding cysteine sites
- Quantitative comparisons (if applicable)
- Bioinformatics interpretation of functional relevance
Can this service support pathway-level or disease-specific studies?
Absolutely. Our data interpretation pipeline includes pathway enrichment, GO annotation, and protein interaction network analysis—ideal for redox signaling, cardiovascular, neurodegenerative, and cancer research models.
Is technical support available after data delivery?
Yes. Our scientific team is available for post-project consultation, helping you interpret complex data, plan follow-up experiments, or publish your results.
Can I use the results from this service for publication?
Yes. All data are generated using validated LC-MS/MS platforms and industry-standard methods. The results are suitable for peer-reviewed publications. If needed, we can provide methodological details and instrument settings for your Materials and Methods section.
Do you offer customized analysis or bioinformatics pipelines?
Absolutely. We understand that research goals vary. Whether you need differential site quantification, pathway clustering, or cross-omics integration, our team can tailor the analysis to meet your specific scientific objectives.
How is the service priced? Are there packages available for multiple samples?
Pricing depends on sample type, number of samples, and selected detection strategy (TMT vs. Biotin Switch). We offer flexible packages for individual projects and discounts for batch or multi-condition studies. Contact us for a customized quote.
Learn about other Q&A.
Case Study

S‐Nitrosoglutathione Reductase Deficiency Causes Aberrant Placental S‐Nitrosylation and Preeclampsia
Journal: Journal of the American Heart Association
Published: 2022Main
doi: 10.1161/JAHA.121.024008
Technology: Botion-HDBP; TMT(Tandem Mass Tag)
- Background
- Results
- Conclusions
Preeclampsia, a leading cause of maternal and fetal mortality and morbidity, is characterized by an increase in S‐nitrosylated proteins and reactive oxygen species, suggesting a pathophysiologic role for dysregulation in nitrosylation and nitrosative stress.
This analysis revealed a markedly increased number of S-nitrosylated residues in GSNOR⁻/⁻ placentas (459 residues corresponding to 351 proteins) compared with controls (264 residues corresponding to 198 proteins) (Figure 1A–1C).
Notably, placentas from ascorbate-treated mice exhibited a higher net number of S-nitrosylated proteins in both control and GSNOR⁻/⁻ groups—consistent with the known transnitrosylation properties of chronic ascorbate treatment—though the increase was less pronounced in the GSNOR⁻/⁻ group (Figure 1D), resulting in a reduced difference in the number of S-nitrosylated proteins between the two groups.
In line with previous findings, the majority of nitrosylated proteins were localized to the cytoplasm and nucleus across all experimental groups (Figure 1E).
To further identify the most affected signaling pathways under conditions of elevated S-nitrosylation, the researchers focused on S-nitrosylated proteins that were uniquely present in GSNOR⁻/⁻ placentas compared to B6 controls and were reversed upon ascorbate treatment. Of all the detected peptides, several S-nitrosylation residues were specific to GSNOR⁻/⁻ placentas, whereas only 16 were unique to B6 (Figures 1C and 1F). All 50 GSNOR⁻/⁻–specific S-nitrosylation residues were reversed by ascorbate. Among the corresponding proteins, 14 have previously been associated with key biological processes relevant to pregnancy, including angiogenesis, inflammation, cell migration, and apoptosis (Figure 1F), underscoring the physiological significance of these findings.
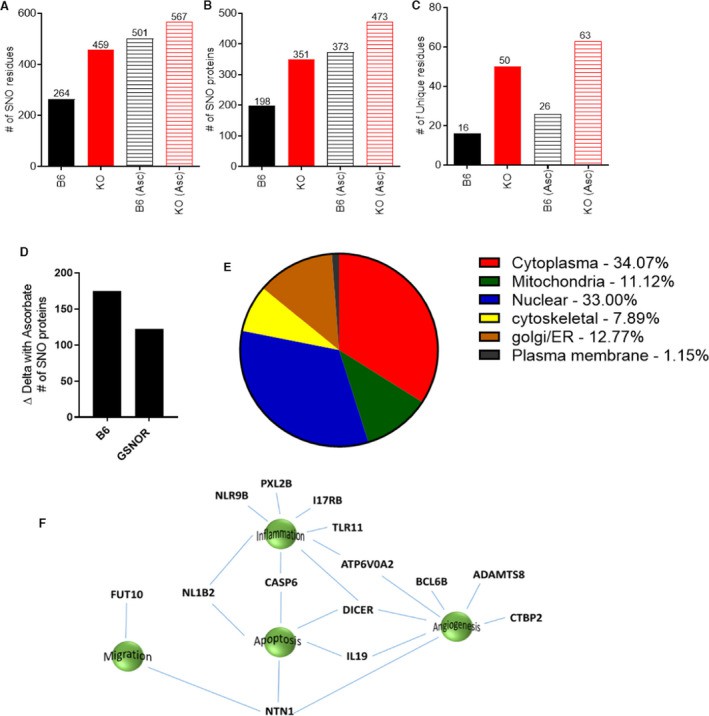 Figure 1: Duel‐labeling mass spectrometry revealed an increased number of SNOylated proteins in the placentas from GSNOR−⁄− animals.
Figure 1: Duel‐labeling mass spectrometry revealed an increased number of SNOylated proteins in the placentas from GSNOR−⁄− animals.
Therefore, deficiency of GSNOR creates dysregulation of placental S‐nitrosylation and preeclampsia in mice, which can be rescued by ascorbate. Coupled with similar findings in human placentas, these findings offer valuable insights and therapeutic implications for preeclampsia.
Publication
Here are some publications in PTMs Proteomics research from our clients:

- Regulation of cardiac ferroptosis in diabetic human heart failure: uncovering molecular pathways and key targets. 2024. Cell Death Discovery. https://doi.org/10.1038/s41420-024-02044-w
- HDAC4 influences the DNA damage response and counteracts senescence by assembling with HDAC1/HDAC2 to control H2BK120 acetylation and homology-directed repair. Nucleic Acids Research. 2024. https://doi.org/10.1093/nar/gkae501
- Synergistic Combined-proteomics Guided Mapping strategy identifies mTOR mediated phosphorylation of LARP1 in nutrient responsiveness and dilated cardiomyopathy. BioRxiv. 2022. https://doi.org/10.1101/2022.10.13.512080
- In Vitro Identification of Phosphorylation Sites on TcPolβ by Protein Kinases TcCK1, TcCK2, TcAUK1, and TcPKC1 and Effect of Phorbol Ester on Activation by TcPKC of TcPolβ in Trypanosoma cruzi Epimastigotes. Microorganisms. 2024. https://doi.org/10.3390/microorganisms12050907
How to place an order:
At Creative Proteomics, many excellent and experienced experts will optimize the experimental protocol according to your requirement and guarantee the high-quality results for protein acetylation. Creative Proteomics provides a broad range of technologies for S-Nitrosylation research that enable quantification of protein amount and S-Nitrosylation modification. Please feel free to contact us by email to discuss your specific needs. Our customer service representatives are available 24 hours a day, from Monday to Sunday.
References
- Sharma V.; et al. S-Nitrosylation in tumor microenvironment. International Journal of Molecular Sciences. 2021; 22(9):4600. DOI: 10.3390/ijms22094600
- Wu X.; et al. Targeting protein modifications in metabolic diseases: molecular mechanisms and targeted therapies. Signal Transduct Target Ther. 2023 May 27;8(1):220. DOI: 10.1038/s41392-023-01439-y
- Falco JA.; et al. Identification of protein targets of S-Nitroso-Coenzyme A-Mediated S-Nitrosation using chemoproteomics. ACS Chem Biol. 2024 Jan 19;19(1):193-207. DOI: 10.1021/acschembio.3c00654










