Integrated Proteomics and Lipidomics Analysis | Multi-Omics Insights
Creative Proteomics offers integrated proteomics and lipidomics analysis to reveal how proteins and lipids interact in complex biological systems. By linking metabolic pathways, molecular interactions, and regulatory mechanisms, our service empowers researchers to achieve deeper systems biology insights, improved biomarker discovery, and publication-ready results.
Submit Your Request Now
×
- Multi-omics integration of proteomics and lipidomics
- Pathway enrichment and biomarker discovery
- Publication-ready deliverables with expert bioinformatics
- What is Integrated Analysis
- Applications
- Core Analyses
- Workflow
- Deliverables
- Advantages
- Sample Requirements
- Case Study
- Publication
Why Integrated Proteomics and Lipidomics Matters
Lipidomics, a vital branch of metabolomics, uncovers how lipid metabolism influences life processes. Proteomics, on the other hand, studies all proteins expressed within an organism, tissue, or cell, analysing their composition and functional patterns at a systems level. Shifts in protein abundance are closely tied to growth, environmental adaptation, and disease progression.
When combined, integrated proteomics and lipidomics analysis creates a powerful framework for multi-omics research. This approach links molecular events across proteins and lipids, providing a more holistic view of cellular regulation. By incorporating functional analysis, pathway enrichment, and molecular interaction mapping, researchers can:
- Explore the regulatory mechanisms of biomolecules at multiple levels.
- Identify key proteins, lipid metabolites, and signalling pathways.
- Build mechanistic models that explain biological changes over time.
Ultimately, this integrated analysis empowers scientists to move beyond descriptive studies. Instead, it enables predictive modelling of molecular networks, supporting deeper biological insights and advancing translational applications in agriculture, biomedicine, and pharmaceutical development.
Applications Across Research Fields
Our integrated multi-omics service supports diverse industries and scientific disciplines:
Agriculture: Plant stress resistance, crop growth, breeding optimisation.
Animal Science: Livestock health, milk and meat quality, disease mechanisms.
Biomedical Research: Biomarkers, disease typing, therapeutic target discovery.
Pharmaceutical R&D: Drug action mechanisms, toxicity evaluation, efficacy studies.
Microbiology: Pathogenesis, antimicrobial resistance, host–pathogen interactions.
Food Science & Nutrition: Functional food development, food safety monitoring, processing optimisation.
What Are the Main Analyses We Provide?
Creative Proteomics integrates proteomic and lipidomic datasets through advanced statistical and bioinformatics approaches. Our workflows are designed to reveal cross-omics interactions, metabolic pathways, and regulatory mechanisms with high confidence.
Core Analyses Offered:
Multivariate Statistical Integration – Combine proteome and lipidome datasets to identify system-wide biological trends.
WGCNA (Weighted Gene Co-Expression Network Analysis) – Detect co-regulated protein–lipid modules linked to key biological processes.
Proteome–Lipidome Correlation Analysis – Map interactions between protein activity and lipid metabolism across conditions.
PCA Comparative Analysis – Apply principal component analysis to distinguish groups and highlight drivers of variation.
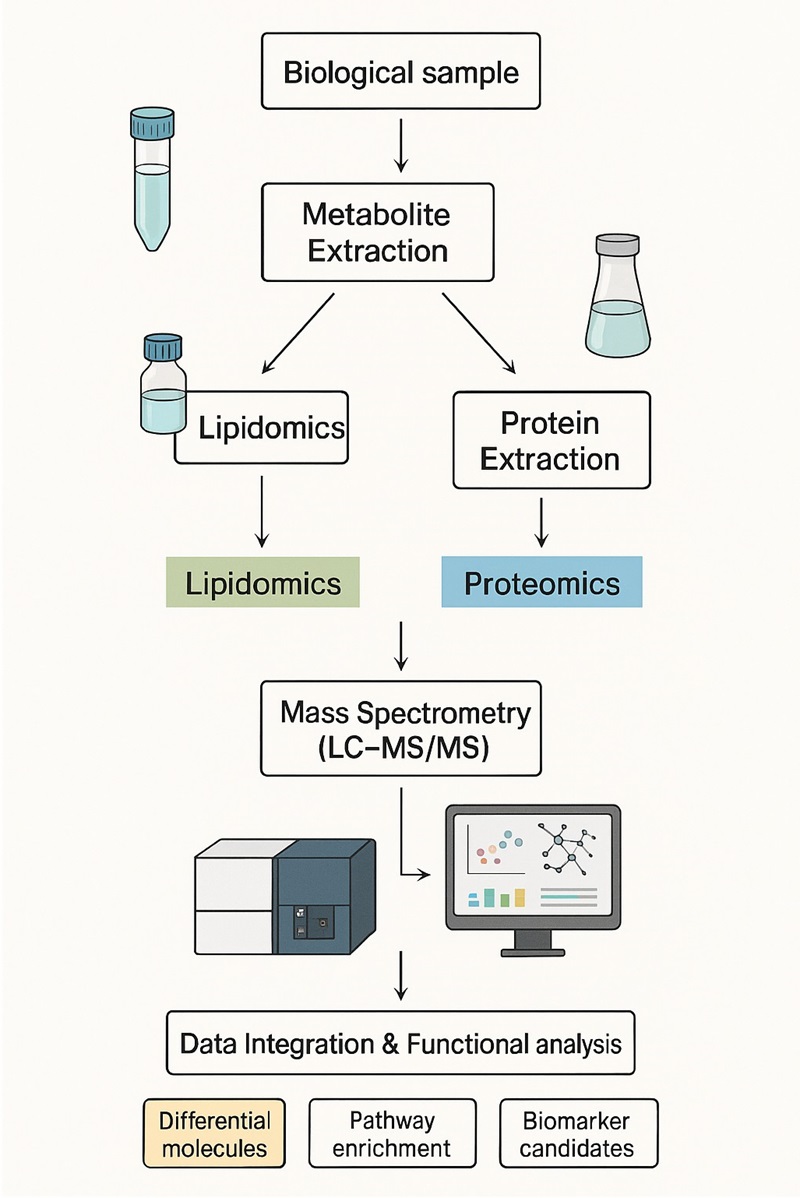
Workflow of Integrated Proteomics and Lipidomics Analysis
Our service combines high-resolution proteomics and lipidomics analysis with advanced bioinformatics integration. Each step is optimised for data quality, reproducibility, and publication-ready outcomes.
1. Sample Preparation
- Extraction of proteins and lipids from biological samples.
- Strict QC procedures ensure integrity and reproducibility.
2. Proteomics Analysis
- High-resolution LC-MS/MS identifies and quantifies proteins.
- Detects changes in expression, modifications, and molecular interactions.
3. Lipidomics Analysis
- Targeted and untargeted LC-MS/MS profiling of lipid species.
- Quantitative coverage of lipid classes and bioactive metabolites.
4. Data Integration
- Multivariate statistics (PCA, WGCNA, correlation networks).
- Pathway enrichment and cross-omics correlation analysis.
5. Bioinformatics Analysis
- Functional annotation and network modelling.
- Identification of biomarkers, regulatory genes, and lipid–protein interaction modules.
6. Reporting & Deliverables
- Comprehensive technical report including QC data, MS images, and bioinformatics outputs.
- Raw data available on request.
- Publication-ready figures (heatmaps, volcano plots, network diagrams).
Deliverables
Comprehensive Technical Report
Detailed experimental procedures, LC-MS/MS parameters, and QC metrics.
Quantitative Datasets
Processed proteomic and lipidomic data tables, including fold-changes and statistical significance.
Raw MS data available upon request.
Data Visualizations
PCA plots, volcano plots, heatmaps, and pathway enrichment diagrams.
Bioinformatics Outputs
Correlation networks, protein–lipid interaction maps, and WGCNA modules.
Interpretative Summary
Expert commentary linking findings to biological pathways and potential biomarkers.
Publication-Ready Figures
Graphs and tables formatted for scientific manuscripts or regulatory submissions.
Advantages of Integrated Proteomics and Lipidomics Analysis
Integrating proteomics and lipidomics delivers a more complete picture of biological systems. Proteomics captures protein expression, modifications, and interactions, while lipidomics reveals lipid composition and dynamics. When combined, these analyses illuminate how proteins and lipids work together to regulate cellular function and metabolism.
Complementary insights emerge by uncovering protein–lipid interactions and post-translational modifications.
Better biomarker discovery happens because integration catches signals that single-omics approaches might miss.
Deeper understanding of disease arises from mapping how proteins and lipids change during pathological progression.
Personalised medicine becomes possible, as integrated molecular profiles can reflect individual variation and support tailor-made interventions.
Sample Requirements for Integrated Proteomics & Lipidomics Analysis
| Sample Type | Proteomics Minimum | Lipidomics Minimum |
|---|---|---|
| Animal tissues | Hard: 300–500 mg Soft: 200 mg | 100–200 mg |
| Plant tissues | Hard: 3–5 g | 100–200 mg |
| Microbial cells | ≥1×10⁷ cells | ≥1×10⁷ cells |
| Cultured cells | ≥1×10⁷ cells (pellet) | ≥1×10⁷ cells |
| Plasma/Serum/CSF | ≥20 µL (undepleted) ≥100 µL (depleted) | ≥100 µL |
| Follicular fluid | 200 µL | N/A |
| Lymph, synovial fluid, ascites | 5 mL | N/A |
| Saliva/Tears/Milk | 3–5 mL | N/A |
| Culture supernatant | 20 mL | N/A |
| Pure protein | 300 µg (in 8 M Urea buffer) | N/A |
| FFPE samples | 15–20 slices (10 µm thick, 1.5×2 cm) | N/A |
Client Case Study

Unlocking Phagocytosis Defects in CLN3 Disease via Integrated Proteomics–Lipidomics Analysis
In a recent study, researchers discovered that impaired phagocytosis of photoreceptor outer segments (POSs) by retinal pigment epithelial (RPE) cells in CLN3 disease is driven by disrupted sphingolipid metabolism. This finding underscores how lipid pathways affect cellular functions critical for vision .
- Our Contribution
- Results and Impact
- Why It Matters
We provided targeted lipidomics profiling (focused on sphingolipid panels) and untargeted proteomics analysis to help unveil the molecular basis of this dysfunction:
- Conducted Targeted Sphingolipid Analysis Panel on CLN3 patient-derived RPE cells to quantify levels of key lipids like S1P.
- Performed Untargeted Proteomic Analysis to assess expression changes in proteins including ASAH1 and Ezrin (EZR).
- Delivered integrated data that correlated lipid level alterations (e.g., decreased S1P) with protein changes that impact RPE microvilli structure and POS phagocytosis.
- We helped reveal that CLN3 disease RPE cells exhibit reduced ASAH1, S1P, and EZR activation—molecular changes linked to defective POS phagocytosis .
- Our analyses supported the mechanism by which supplementing recombinant ASAH1 restored S1P levels, re-activated EZR, and rescued phagocytosis functionality.
This integrated proteomics–lipidomics case provided both lipid and protein-level evidence to explain RPE dysfunction in CLN3 disease. Our work enabled:
- Identification of a metabolic–structural disease axis (ASAH1–S1P–EZR) in RPE cells.
- Insights that could guide targeted therapeutic strategies, such as ASAH1 supplementation or modulation of sphingolipid pathways.
Publication

- Kumar LK, Han J, Dalvi S, et al. (2024) "Impaired phagocytosis of photoreceptor outer segments by RPE in CLN3 disease is a consequence of altered sphingolipid metabolism." Investigative Ophthalmology & Visual Science, 65(7):1565.
- Foley N, Dalvi S, Kumar LK, et al. (2024) "Cell-autonomous RPE dysfunction is sufficient to promote retina degeneration in an iPSC-based CLN3 disease model." Investigative Ophthalmology & Visual Science, 65(7):1563.
- Han J, Chear S, Talbot J, et al. (2024) "Genetic and cellular basis of impaired phagocytosis and photoreceptor degeneration in CLN3 disease." bioRxiv preprint.
References
- Liu H,Hong Y,Lu Q, et al. Integrated Analysis of Comparative Lipidomics and Proteomics Reveals the Dynamic Changes of Lipid Molecular Species in High-Oleic Acid Peanut Seed. J Agric Food Chem. 2020;68 (1):426-438.
- Madrid-Gambin F, Föcking M, Sabherwal S, Heurich M, English JA, O'Gorman A, Suvitaival T, Ahonen L, et al. Integrated Lipidomics and Proteomics Point to Early Blood-Based Changes in Childhood Preceding Later Development of Psychotic Experiences: Evidence From the Avon Longitudinal Study of Parents and Children. Biol Psychiatry. 2019 Jul 1;86(1):25-34. doi: 10.1016/j.biopsych.2019.01.018. Epub 2019 Jan 30. PMID: 30878195; PMCID: PMC6579334.












