Protein primary structure is the linear amino acid sequence that makes up the protein polypeptide chain. Protein amino acid sequence analysis is the foundation and key of protein research. The information obtained by protein sequencing has many valuable uses, such as the identification of proteins, the design of probes for molecular cloning, and the synthesis of peptides for immunogens. Protein sequencing also plays a crucial role in understanding the structure, function and interactions of peptides or proteins. Accurate determination of sequence allows researchers to reveal important information about biological processes, identify post-translational modifications, and develop targeted therapies.
Comprehensive One-Stop Protein Sequence Analysis Service
Creative Proteomics utilizes nano LC-MS/MS and Edman degradation sequencing systems to analyze protein sequences. This includes analysis of the amino acid composition of proteins, N-terminal sequencing, C-terminal sequencing, full sequence analysis, as well as N-terminal sequence analysis services based on Edman degradation. For proteins with unknown theoretical sequences, Creative Proteomics offers protein de novo sequencing services to analyze protein sequences.
N-terminal Edman Degradation Service:
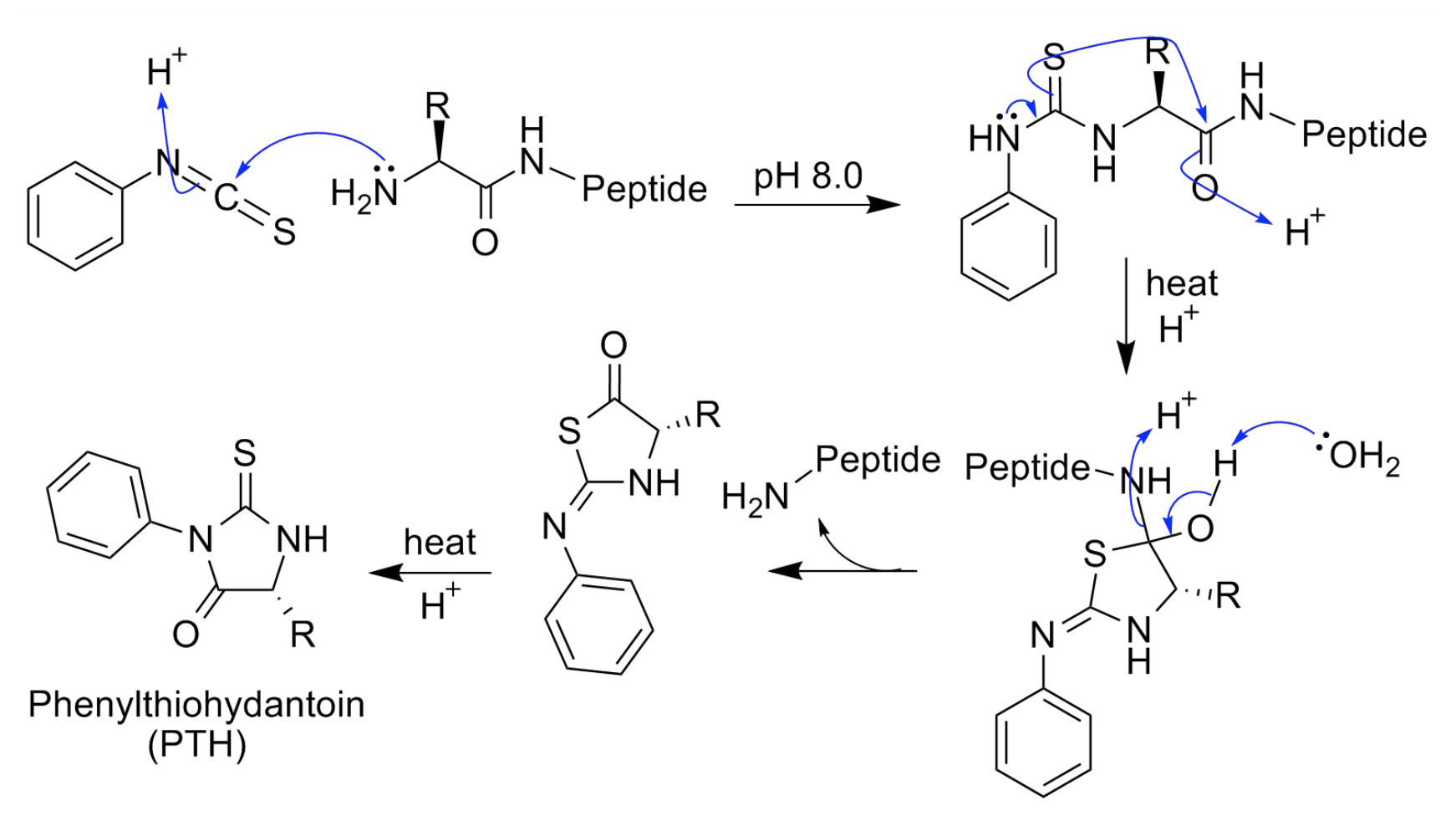
In protein N-terminal sequence analysis, proteins are first modified with phenylisothiocyanate (PITC). Subsequently, the derivatized terminal amino acid is removed by acid cleavage in a form of phenylthiohydantoin (PTH) derivative and a new α-amino group on the next amino acid is now available for the next round of reaction with PITC. Protein sequence is therefore being analyzed in through a serious reaction of PITC addition, and cleavage of one PTH at each time for analysis. Suppose the reactions are fully efficient, this method can sequence the whole amino acid from the N-terminal end. Though less efficient as compared with mass spectrometer, N-terminal Edman sequencing still has advantages for protein analysis that cannot readily be obtained by other analysis methods. With current technology, it is fairly routine to obtain at least 20 to 25 residues of sequence from the N-terminus of the proteins and peptides.
Mass spectrometry:
Currently, the most commonly used technique for protein sequence analysis is mass spectrometry. Mass spectrometry is a highly efficient method for the accurate mass determination and characterization of proteins. Two mainly used mass spectrometers are electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI), respectively. Basically, proteins are pretreated with digestive enzymes to be digested into small fragments, which are analyzed by mass spectrometers. Depends on the projects need, MALDI and ESI are two used as ionization source for ionizing peptide fragments. Usually, MALDI is more frequently used when dealing with large amount of samples. Peptide sequences are then obtained by analyzing the mass spectrum of each of the fragments, which together consist of the full-length protein sequence.
Protein De Novo Sequencing
The biggest feature of the protein de novo sequencing method is that it can directly analyze the protein sequence without using any protein or DNA database information. Compared with the traditional mass spectrometry database search method, it has an incomparable advantage, that is, the de novo sequencing method can be applied to analyze the protein sequences of new species, as well as the protein sequences of species whose genomes have not been sequenced.
De Novo Antibody Sequencing
Antibodies play a vital role in immune response and targeted therapy. De novo antibody sequencing allows the determination of the complete amino acid sequence of an antibody, including both variable and constant regions. This information is crucial for antibody engineering, therapeutic antibody development, and understanding the immune response. Creative Proteomics specializes in de novo antibody sequencing services, utilizing advanced techniques such as liquid chromatography-mass spectrometry (LC-MS) and bioinformatics tools to accurately sequence and characterize antibodies. By providing comprehensive antibody sequence information, Creative Proteomics facilitates antibody optimization and development of antibody-based therapeutics.
C-Terminal Sequencing
In addition to N-terminal sequencing, determining the C-terminal sequence of a protein is equally important for understanding its structure and function. Creative Proteomics offers C-terminal sequencing services that enable researchers to precisely identify the amino acids at the C-terminus of a protein or peptide. Various techniques, such as carboxypeptidase digestion or matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), are employed to accurately determine the C-terminal sequence. By incorporating C-terminal sequencing into their repertoire of services, Creative Proteomics assists researchers in gaining comprehensive insights into protein structures and post-translational modifications.
Application of Protein Sequencing Service
Protein Structure Determination
One of the key applications of protein sequencing services is in the determination of protein structures. The amino acid sequence provides the foundation for predicting the secondary and tertiary structures of proteins. By knowing the sequence, researchers can make predictions about protein folding patterns, identify potential binding sites, and gain insights into the protein's overall architecture. Creative Proteomics, with its expertise in protein sequencing, aids researchers in unraveling the complex structures of proteins and advancing our understanding of their functions.
Post-Translational Modification Analysis
Post-translational modifications (PTMs) play a crucial role in regulating protein function and cellular processes. They can include phosphorylation, glycosylation, acetylation, and many other modifications that alter the chemical properties and activity of proteins. Protein sequencing services offered by Creative Proteomics enable the identification and characterization of PTMs. By analyzing the sequence and detecting modifications at specific residues, researchers can uncover the intricate regulatory mechanisms involved in protein signaling, metabolism, and disease pathways.
Proteomic Profiling
Protein sequencing services also contribute to proteomic profiling studies, where researchers aim to identify and quantify the proteins present in a biological sample. By combining techniques like mass spectrometry-based protein sequencing with advanced data analysis, researchers can create comprehensive catalogs of proteins in various biological systems. These catalogs provide valuable insights into cellular processes, biomarker discovery, and disease mechanisms. Creative Proteomics offers proteomic profiling services, empowering researchers to delve deeper into the proteome and unravel its complexities.
Drug Development and Therapeutics
Protein sequencing plays a significant role in drug development and the field of therapeutics. Understanding the precise sequence of therapeutic proteins, such as antibodies or recombinant proteins, is essential for ensuring their safety, efficacy, and proper functionality. Creative Proteomics provides protein sequencing services that aid in quality control and verification of therapeutic proteins, helping to bring safe and effective drugs to market. Additionally, de novo sequencing services offered by Creative Proteomics contribute to the discovery and development of novel therapeutic targets, facilitating the design of innovative therapies.
Here are more related services on protein sequence analysis that you may be interested in:
Protein N-terminal sequencing based on Edman degradation
Protein/Peptide de novo sequencing
Protein sequencing based on top-down approach
Sequence analysis based on mass spectrometry
Technology platform:
- Liquid Chromatography (LC)
- High Performance Liquid Chromatography (HPLC)
- Matrix Assisted Laser Desorption Ionization Mass Spectrometry (MALDI-MS)
- Nano HPLC- 7 T solariX XR FTICR-MS (equipped with ECD)
- Nano HPLC- Orbitrap Fusion™ Lumos™ Tribrid™ MS (equipped with ETD)
Ordering Procedure:







