Cell Surface Proteomics
Creative Proteomics offers advanced solutions for the identification, quantification, and functional analysis of cell surface proteins (CSPs). Utilizing cutting-edge LC-MS/MS-based quantitative proteomics techniques, we provide high-throughput, highly accurate, and specific analysis of complex protein mixtures. Our services include CSP enrichment through methods like differential and density gradient centrifugation, biotinylation, and chemical affinity strategies, as well as robust proteomic quantification techniques such as SILAC, iTRAQ/TMT, label-free, and targeted proteomics.
Submit Your Request Now
×- Define
- What We Provide
- Advantages
- Sample Requirements
- Demo
- Case
- FAQs
- Publications
What is Cell Surface Proteins?
The cell-surface proteins (CSPs) are a group of proteins that traverse or are anchored/embedded in the plasma membrane, playing a crucial role in various essential and distinct cellular functions along the plasma membrane, which mediates intercellular communication and regulating cellular interactions with the extracellular milieu, including nutrient and ion transportation, intercellular interactions, receptor-mediated signaling transduction, enzymatic reactions, and immune recognition. According to Almen et al., they classified CSPs into four categories: (i) transporters, including channels, solute carriers, and active transporters; (ii) receptors, such as G protein-coupled receptors, receptor type tyrosine kinases, receptors of the immunoglobulin superfamily and related, and scavenger receptors and related; (iii) enzymes, which include oxidoreductases, transferases, hydrolases, lyases, isomerases and ligases; and (iv) miscellaneous, such as ligands, other proteins, structural/adhesion proteins and proteins of unknown function. Due to their critical biological function and unique subcellular location, CSPs have been proposed as valuable resource for the identification of targets in immune and targeted therapy. Additionally, they have been served as informative biomarkers for assays related to early disease detection, diagnosis, and prediction. Therefore, it is essential to employ cell surface proteomics strategies for identifying and quantifying the complete repertoire of CSPs, as well as investigating their functional roles.
Our Cell Surface Proteomics Service
With advancements in quantitative proteomics and shotgun proteomics based on LC-MS/MS analysis, it is now capable of characterizing and quantifying complex protein mixtures with high throughput, exclusive accuracy, and specificity. The initial step in studying CSPs involves the isolation of these proteins from the whole cells. Various strategies can be employed for the enrichment of CSPs, including (i) differential centrifugation, (ii) density gradient centrifugation, (iii) affinity-based enrichment methods such as streptavidin-biotinylation, antibody-antigen, and lectin-glycan, and (iv) hydrazide chemistry. Combining centrifugation with affinity or chemical enrichment techniques is a favorable approach to effectively remove abundant cytosolic proteins and enhance selectivity towards CSPs. The second critical step is to dissolve the CSPs, usually using agents such as detergents (SDS, RapiGest, etc.), denaturants (Urea, etc.), extreme pH (Na2CO3, etc.), high ionic strength (KCl, NaCl, etc.), organics (CH3OH, CHCl3, etc.) to dissolution CSPs. The proteomics technology is undoubtedly the most crucial step for deep and accurate identification and quantification of CSPs. The field of quantitative proteomics has witnessed the establishment of various well-established techniques, including SILAC, iTRAQ/TMT, label-free, DIA and target quantification proteomics such as (SRM/MRM, PRM). Most importantly, the methods mentioned above have been thoroughly mastered by Creative Proteomics. With over a decade of experience in the field of proteomics, we possess exceptional proficiency in executing Cell Surface Proteomics Projects. We are capable of supporting diverse sample types and can even assist in cell culturing for SILAC quantitative proteomics.
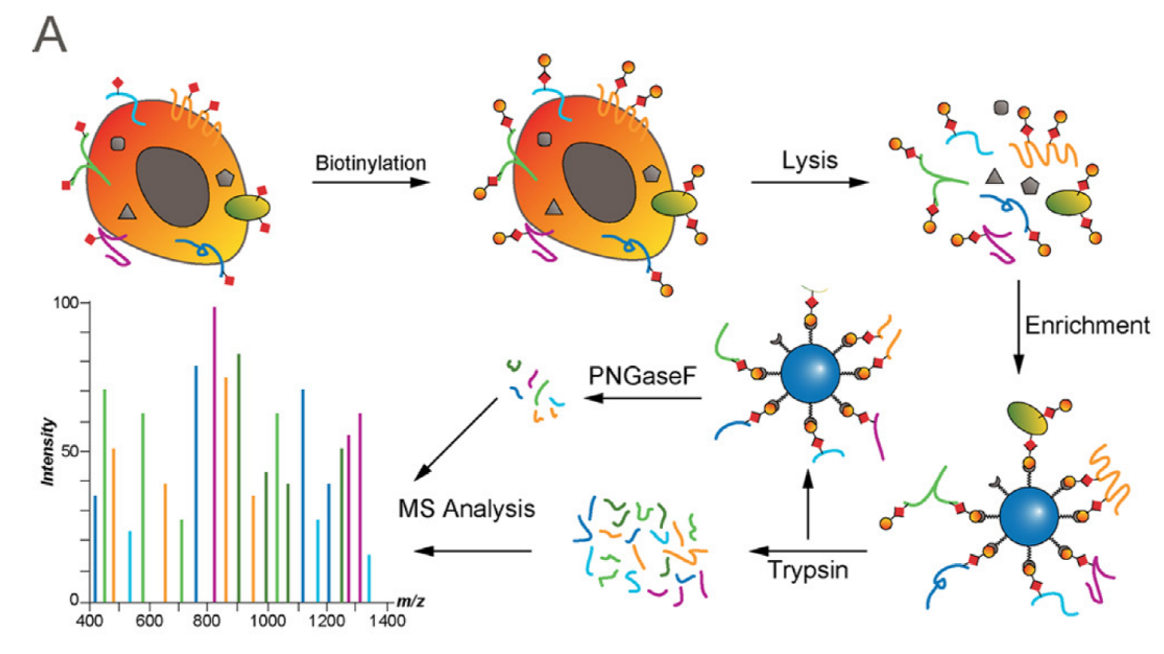 Figure 1. Biotinylation enrichment methods for cell surface proteomics (Kalxdorf M. Molecular Cell Proteomics. 2017).
Figure 1. Biotinylation enrichment methods for cell surface proteomics (Kalxdorf M. Molecular Cell Proteomics. 2017).
Advantages of Cell Surface Proteomics Service
- Professional detection and analysis capability: Experienced technical team, strict quality control system, together with ultra-high resolution detection system and professional data pre-processing and analysis capability, ensure reliable and accurate data.
- High specificity and purification: Optimization of experimental design and methods.
- High stability and reproducible: Obtain consistent and reproducible inter- and intra- assay results for data analysis.
- High throughput: LC-MS/MS can identify and quantify thousands of proteins simultaneously.
- High resolution and sensitivity: Triple TOF 5600, Q-Exactive, Q-Exactive HF, Orbitrap Fusion™ Tribrid™, etc.
- High selectivity: We can provide a wide range of multi-technological services and efficiently handle various types of samples, while remaining cost-effective and ensuring short turnaround times for your projects.
Sample Requirements for Cell Surface Proteomics
| Sample Type | Description | Recommended Sample Amount |
|---|---|---|
| Adherent Cell Lines | Cultured cells that attach to the substrate. | 1 x 106 cells (approx. 0.1 g) |
| Suspension Cell Lines | Cultured cells that do not attach to the substrate. | 1 x 107 cells (approx. 0.5 g) |
| Primary Cells | Isolated from tissues; can be adherent or suspension. | 1 x 106 to 1 x 107 cells |
| Tissue Samples | Fresh or frozen tissue samples (e.g., liver, muscle). | 100-500 mg |
| Plasma/Serum Samples | Liquid biopsies from blood; may contain cell surface proteins. | 1-5 mL |
| Biopsies | Small tissue samples taken for diagnostic purposes. | 50-100 mg |
| Microbial Cells | Bacterial or fungal cultures for surface protein study. | 1 x 108 cells |
| Cell Membrane Fractions | Isolated membrane fractions from any cell type. | 50-100 mg |
| Exosomes | Small vesicles from cell cultures or biological fluids. | 10-50 mL of source fluid |
| Organelles | Isolated organelles such as membranes or cytoplasmic vesicles. | 50-100 mg |
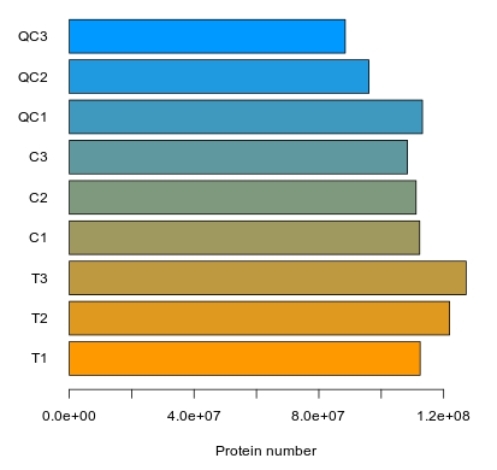
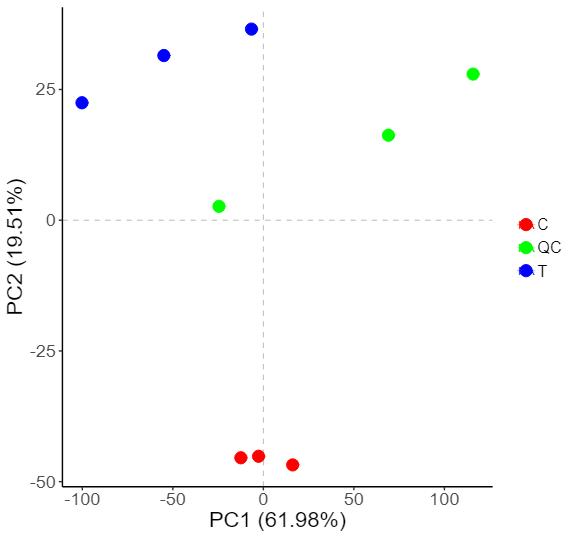
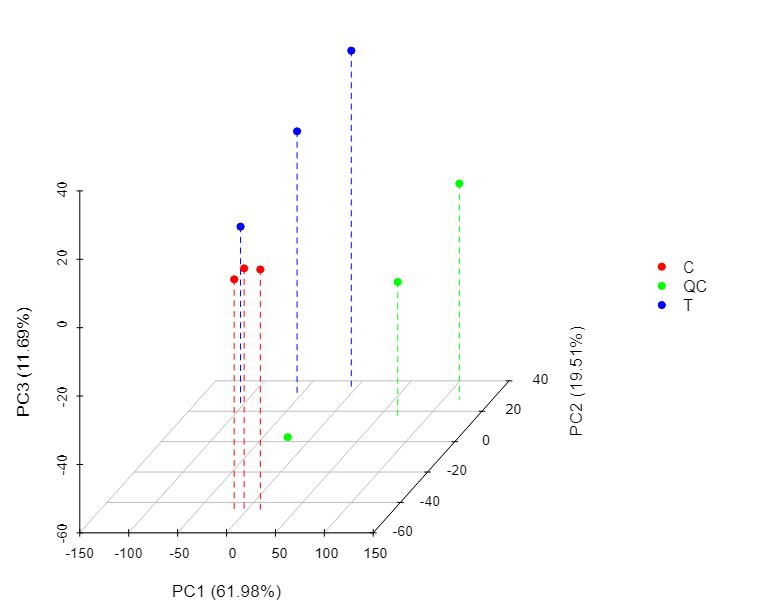
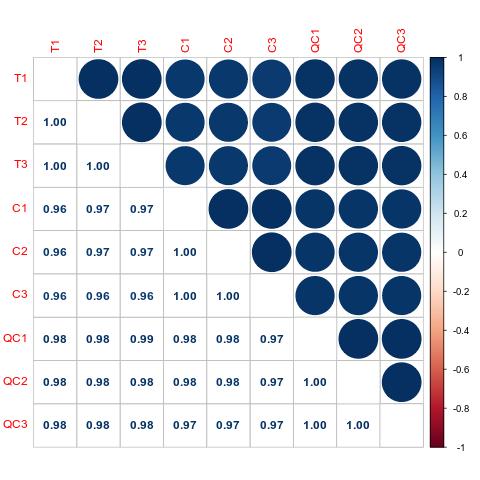
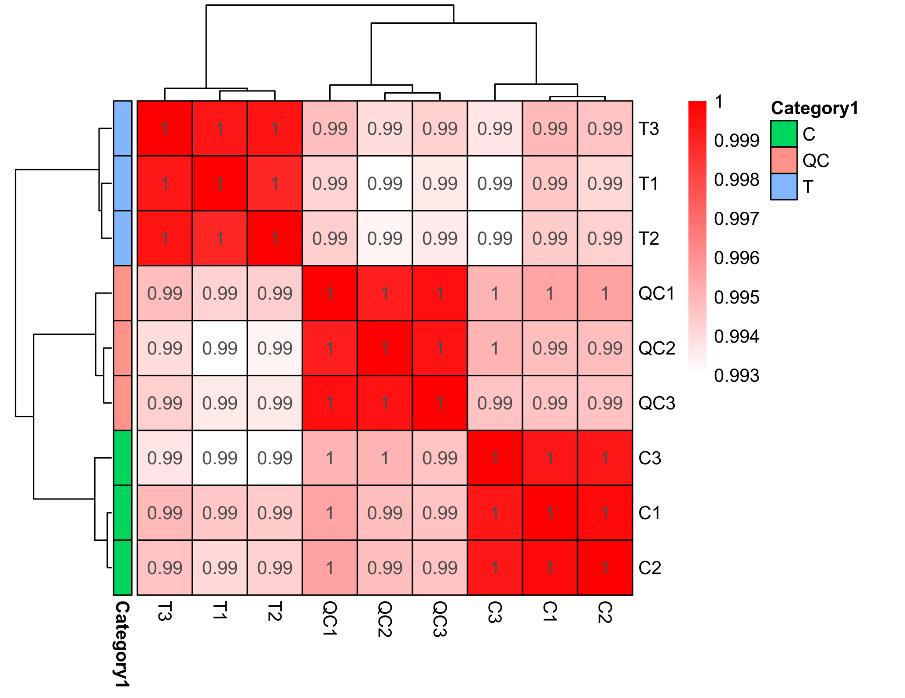
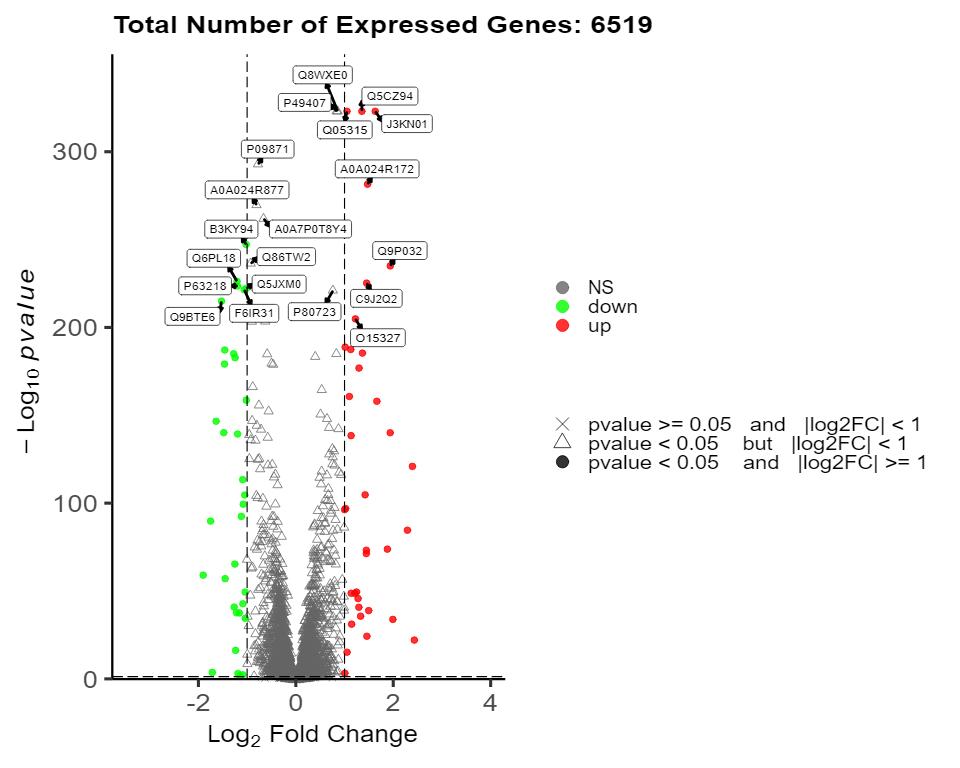
Demo
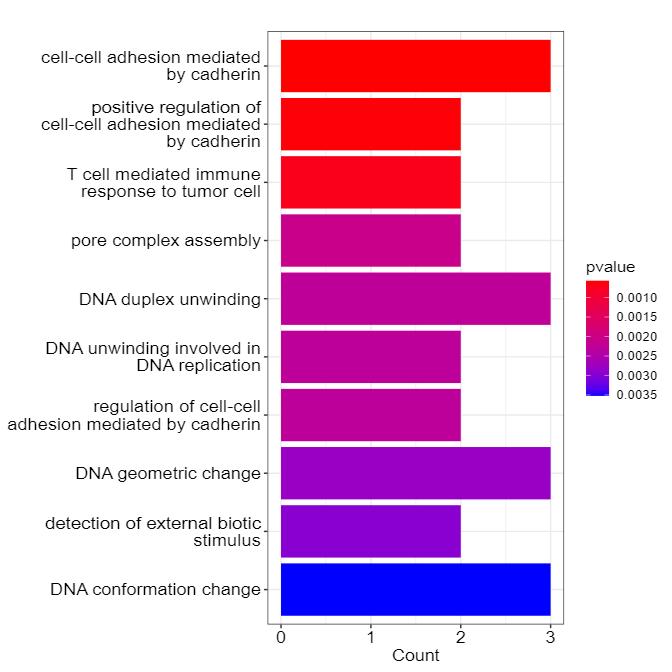
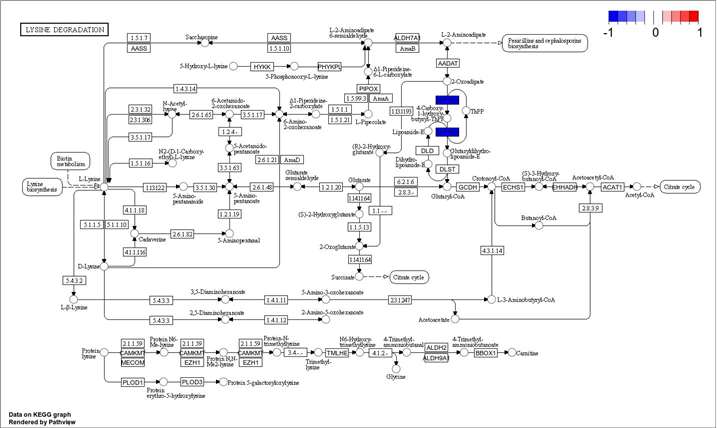
Case Study
FAQ for Cell Surface Proteomics
How are cell surface proteins isolated from other cellular components?
Isolation of cell surface proteins (CSPs) involves several enrichment strategies. Common methods include differential centrifugation to separate membrane proteins from cytosolic proteins, density gradient centrifugation, and affinity-based techniques such as biotinylation followed by streptavidin capture. Each method has its advantages, and often, a combination of techniques is employed to enhance selectivity for CSPs while reducing the presence of abundant cytosolic proteins.
How do you handle the data analysis for cell surface proteomics?
Data analysis in cell surface proteomics is conducted using a combination of bioinformatics tools and statistical approaches. After raw data acquisition via LC-MS/MS, we perform rigorous data processing, including peak identification and quantification. Our analysis pipeline includes bioinformatics applications for downstream analysis such as protein-protein interaction networks, pathway analysis (e.g., KEGG, GO), and comparative analysis between different sample groups. Detailed reports are generated to summarize the findings, and we also offer custom analysis options based on client needs.
What are the potential applications of cell surface proteomics in biomedical research?
Cell surface proteomics has a wide array of applications in biomedical research, including:
- Identifying potential biomarkers for early disease detection and diagnosis.
- Studying immune responses and the interactions of immune cells with pathogens.
- Investigating tumor microenvironments and the roles of CSPs in cancer progression.
- Understanding cellular responses to therapeutic agents and drug resistance mechanisms.
- Exploring plant-microbe interactions in agricultural research.
What precautions should be taken regarding sample composition before submission?
It is crucial to ensure that submitted samples are free from nucleic acids, lipids, polysaccharides, detergents, and denaturants, as these components can interfere with protein extraction and analysis. We recommend informing our team about the buffer composition used in protein solutions to allow for proper processing and to avoid any adverse reactions that could affect the integrity of the analysis.
Learn about other Q&A about proteomics technology.
Publications
Here are some publications from our clients:

- Quantitative proteomic analysis of cellular responses to a designed amino acid feed in a monoclonal antibody producing Chinese hamster ovary cell line. 2018. https://doi.org/10.29252/.22.6.385
- Variability in probiotic formulations revealed by proteomics and physico-chemistry approach in relation to the gut permeability. 2020. https://doi.org/10.1007/s12602-019-09590-1
- Untargeted proteomics and stage-specific Huntington's disease reveal biological pathways, and potential protein biomarkers. 2024. https://doi.org/10.21203/rs.3.rs-4508811/v1
- Impaired phagocytosis of photoreceptor outer segments by RPE in CLN3 disease is a consequence of altered sphingolipid metabolism. 2024. [DOI not provided]