- Service Details
- Case Study
What are Eicosanoids and their functions?
Eicosanoids include prostaglandins (PG), thromboxanes (TX), leukotrienes (LT), and lipoxins (LX). Prostanoids refer to PGs and TXs. The names of prostanoids are given according to the number of carbon-carbon double bonds in the molecule. Because of the presence of two carbon-carbon double bonds, most of the biologically active prostaglandins and thromboxanes are series 2 molecules. The eicosanoids with four carbon-carbon double bonds are series 4 molecules. Prostaglandins are first shown being produced in the prostate gland. Thromboxanes are first shown being synthesized in platelets (thrombocytes). And leukotrienes are first identified from leukocytes. Therefore, they were given their respective names. The lipoxins are inflammation involved with eicosanoids, which are produced through lipoxygenase interactions. In response to ingestion of aspirin, the synthesis of lipoxins can be increased according. Therefore, lipoxins are powerful inflammation modulating eicosanoid compounds. Other eicosanoid derivatives include resolvins (Rv) and the protectins (PD).
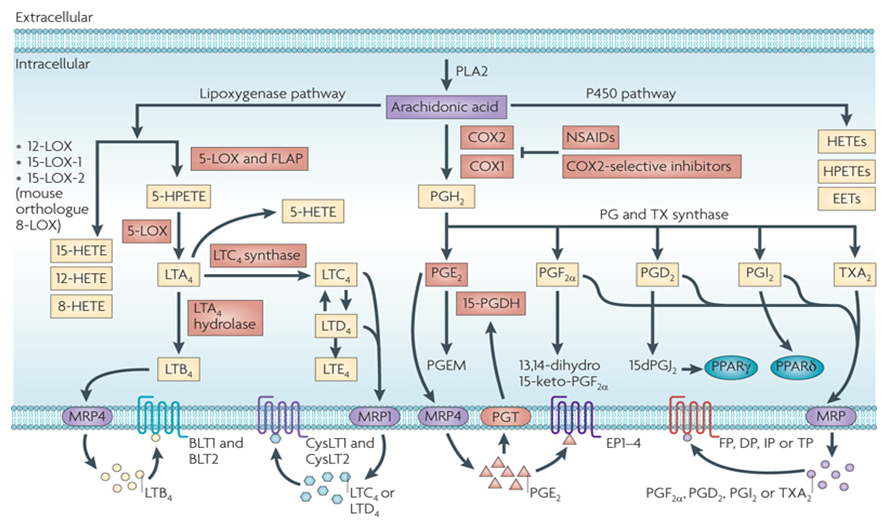
Eicosanoids play an important role in the inflammatory responses of the joints, skin and eyes, on the induction of labor and on the intensity and duration of pain and fever. They also assist in the inhibition gastric acid secretion, regulation of blood pressure through vasodilation or constriction, and inhibition or activation of platelet aggregation and thrombosis. The eicosanoids of greatest biological importance to humans are a large number of molecules arachidonic acid derivatives in the reaction pathway from linoleic acid to arachidonic acid. There is also a small group of eicosanoids comes from eicosapentaenoic acid derived from α-linolenic acid. The main source of arachidonic acid is cellular stores release. Inside the cell, arachidonic acid localized primarily at the C–2 positions of membrane phospholipids and the release is triggered by the activation of PLA2.
Eicosanoids can be synthesized in all mammalian cells except erythrocytes. These molecules are rather powerful, minimum amount of eicosanoids can exert potent physiological effects. At the site of synthesis, eicosanoids exert their roles locally through receptor-mediated G-protein linked signaling pathways. There are two main pathways are associated with the biosynthesis of eicosanoids. Prostaglandins and thromboxanes are generated in the cyclic pathway, while the leukotrienes are produced in the linear pathway.
Eicosanoid Quantitative Analysis Services
Since eicosanoids play a crucial role in large number of disorders like asthma, cardiovascular disease, cancer and chronic obstructive pulmonary disease, the detection and quantification of these compounds are of great interest. Since the endogenous eicosanoids are of rather low concentration, sensitive and specific analytical methods are required for the reliable quantification of these compounds. HPLC-MS/MS has emerged as one of the main techniques used for eicosanoid quantification. Creative Proteomics has established sensitive, reliable, and accurate LC-MS/MS method for quantification of eicosanoids.
Platform
- LC-MS/MS
Summary
- Identification and quantification of eicosanoids by LC-MS/MS.
Technical Advantages
Platform advantages: orbitrap mass analyzer, ultra-high resolution mass spectrometry, high-quality data
Wide applicability: no species restriction, no standard product restriction
High throughput: detect dozens of lipid molecules at one time, saving samples and costs
Strong quantitative ability: the sensitivity can reach ppm level, and the linear range can reach 5-6 orders of magnitude
State of art facilities
Constantly optimized protocol and analytical software
Professional experiment design
Quick turnaround time
Sample requirement
| Sample Type | Minimum Sample Amount |
|---|---|
| Cells | Not less than 1x10^7 cells |
| Tissues | Not less than 250 mg |
| Serum, Plasma | Not less than 300 μL |
| Feces and Intestinal Contents | Not less than 200 mg |
| Microorganisms (Bacteria, Fungi, etc.) | Not less than 500 mg, not less than 10^7 CFU/mL |
| Other Liquid Samples | "Urine not less than 1 mL, Saliva not less than 500 μL, |
| Amniotic Fluid, Bile, Cerebrospinal Fluid, Lymph Fluid, etc., not less than 300 μL" |
Report
- A detailed technical report will be provided at the end of the whole project, including the experiment procedure, GC-MS instrument parameters
- Analytes are reported as uM or ug/mg (tissue), and CV's are generally<10%
- The name of the analytes, abbreviation, formula, molecular weight and CAS# would also be included in the report.
| Eicosanoids Quantified in This Service | ||
|---|---|---|
| (±)10-HDoHE | (±)11,12-DHET | (±)11,12-EET |
| (±)11-HDoHE | (±)11-HETE | (±)12(13)-EpOME |
| (±)12-HETE | (±)13-HDoHE | (±)13-HODE |
| (±)14,15-DHET | (±)14,15-EET | (±)16-HDoHE |
| (±)16-HETE | (±)17-HDoHE | (±)17-HDoHE |
| (±)17-HETE | (±)18-HETE | (±)20-HDoHE |
| (±)20-HETE | (±)4-HDoHE | (±)5,6-DHET |
| (±)5-HETE | (±)7-HDoHE | (±)8,9-DHET |
| (±)8,9-EET | (±)8-HDoHE | (±)9(10)-EpOME |
| (±)9-HETE | 11,12-EEQ | 11-dehydro TXB2 |
| 11-dehydro TXB3 | 11-deoxy PGF2α | 11-HEPE |
| 12-HEPE | 12-keto-LTB4/12-oxo-LTB4 | 12-OxoETE |
| 13,14-dihydro PGE1 | 13,14-dihydro PGF1α | 13,14-dihydro-15-keto PGA2 |
| 13,14-dihydro-15-keto PGE1 | 13,14-dihydro-15-keto PGF1α | 13,14-dihydro-15-keto PGF2α |
| 13,14-dihydro-15-keto Prostaglandin D1 | 13,14-dihydro-15-keto Prostaglandin D2 | 13,14-dihydro-15-keto Prostaglandin E2 |
| 13-OxoETE | 13-OxoODE | 14 15 ,15-EEQ |
| 14,15-DiHETE | 15 (S)--HPETE | 15/6-keto-PGF1α |
| 15-deoxy-Δ12,14-Prostaglandin D2 | 15-deoxy-Δ12,14-Prostaglandin J2 | 15-HEPE |
| 15-HETE | 15-keto PGA1 | 15-keto PGE1 |
| 15-keto PGE2 | 15-Keto-PGF2a | 16,17-EDP |
| 17,18-DiHETE | 17,18-EEQ | 18-HEPE |
| 19(R)-HETE | 19(R)-hydroxy prostaglandin E2 | 19(R)-hydroxy prostaglandin F2a |
| 19,20-EDP | 20-carboxy LTB4 | 20-OH-LTB4 |
| 5 (S)-HpETE | 5(6)-EET | 5(S),14(R)-Lipoxin B4 |
| 5(S),6(R)-DiHETE | 5(S),6(R)-Lipoxin A4 | 5,6-DiHETE |
| 5-HEPE | 5-OxoETE | 6-keto PGE1 |
| 6-keto Prostaglandin F1α | 6-trans leukotriene E4 | 8,9-EEQ |
| 8-HEPE | 8-HETE | 8-iso PGE2 |
| 8-iso PGF2α | 8-iso Prostaglandin A1 | 8-iso Prostaglandin A2 |
| 8-iso-15-keto PGE2 | 8-iso-prostane | 9-HEPE |
| 9-HETE | 9-HODE | 9-OxoODE |
| Arachidonic acid | Docosahexaenoic Acid | Docosapentaenoic Acid |
| Eicosapentaenoic Acid | lecithin | Leukotriene B4 |
| LTB4 | LTC4 | LTD4 |
| LTE4 | Maresin-1 | N-acetyl leukotriene E4 |
| PGA2 | PGE1 | PGF1α |
| PGF2α | PGF3α | PGG2 |
| PGH2 | PGJ2 | Prostaglandin B2 |
| Prostaglandin D2 | Prostaglandin D3 | Prostaglandin E2 |
| Prostaglandin E3 | Resolvin-D1 | Resolvin-D2 |
| Thromboxane B2 | TXB3 | Δ12-PGD2 |
| Δ12-PGJ2 | ||
Ordering Procedure:

With integrated set of separation, characterization, identification and quantification systems featured with excellent robustness & reproducibility, high and ultra-sensitivity, Creative Proteomics provides reliable, rapid and cost-effective eicosanoids targeted lipidomics services.
Phospholipids and lysophosphatidic acids in relation to ulcerative colitis
Lipidomic Trajectories Characterize Delayed Mucosal Wound Healing in Quiescent Ulcerative Colitis and Identify Potential Novel Therapeutic Targets
Journal: International Journal of Biological Sciences
Published: 2022
Abstract:
In previous studies, only a handful of lipids have been revealed to play a role in human wound healing. However, due to the lack of suitable human models, research on phospholipids and class-specific fatty acids in human colonic mucosal wound healing remains underexplored. This study utilized ultra-high-performance liquid chromatography-mass spectrometry (UPLC-MS) to analyze the composition of phospholipids and class-specific fatty acids in two cohorts. Several phospholipids and class-specific fatty acids, particularly LPC, PC, LPA-PG, PI, PGD2, and PGE1, were identified in inflamed colonic tissues, representing a potential novel therapeutic approach for restoring inflammation in UC and subsequent epithelial healing, revealing new treatment targets for ulcerative colitis.
Methods: Targeted lipidomics
Ultra-high-performance liquid chromatography-mass spectrometry (UPLC-MS) was used to identify phospholipids and class-specific fatty acids. Clinical tissue samples were weighed and extracted with internal standards, BHT, and methanol. Samples were separated using a reverse-phase C18 column, detected under electrospray ionization (ESI) mode, and data were collected in multiple reaction monitoring (MRM) mode. Negative ion mode quantified some phospholipids and class-specific fatty acids, while positive ion mode quantified other types of phospholipids.
Results and Discussion:
A total of 98 phospholipids were detected and identified. Statistical analysis revealed significant differences in all detected phospholipid categories between day 0 and days 2 and 7 in the UC group, while there were no significant differences between day 0 and day 7 in the control group. A total of 33 class-specific fatty acids were detected and identified. It was observed that, compared to day 0, all groups showed significant reductions on days 2 and 7, with some being specific to UC, such as PGE1, 13,14-2OH-15keto-PGD2, PGJ2, 5(S)-HEPE, 13-HODE, and 14,15-di-HETE, while others were specific to the control group, such as 5(S)-HETE, 8-HETE, and 5(6)-EET. However, no differences were found when comparing UC and control groups at the three time points.

Based on the phospholipid OPLS-DA model, significant changes were observed between the UC and control groups, particularly with a higher number of affected phospholipids in the UC group (p < 0.05). On day 7, significant differences were noted between the UC and control groups, with various phospholipid classes, including phosphatidylserine (PS), phosphatidylinositol (PI), phosphatidylglycerol (PG), phosphatidylcholine (PC), phosphatidic acid (PA), and lysophosphatidic acid (LPA), all showing a significant decrease in levels within the UC group.


Based on the OPLS-DA model for classifying lysophosphatidic acids (LPAs), it was observed that the UC group had a greater number of significantly affected class-20 sulfonic acids compared to the control group (p < 0.05). However, there were no significant differences between the UC and control groups. Significant temporal variations in the levels of class-20 sulfonic acids were discovered during the injury and wound healing process, but there were no significant differences in metabolic trajectories between the UC and control groups.

Finally, based on the OPLS-DA model, a correlation analysis was conducted between wound scores (x-matrix) and phospholipids (y-matrix). The wound scores showed a high correlation with the phospholipid profile, especially with changes in sphingomyelins (SM) and phosphatidylethanolamines (PE), which were positively correlated with wound scores. In contrast, the levels of phosphatidylglycerol (PG) and lysophosphatidic acids (LPA) were negatively correlated with wound scores.

This study provides the first dynamic description of metabolic pathway disturbances during colonic mucosal wound healing in healthy individuals and quiescent UC patients. The analysis identified sustained inflammatory responses and delayed epithelial recovery during UC wound healing, along with UC-specific perturbations involving lipid mediators. Significant changes over time were observed in several phospholipids and arachidonic acid metabolites. Therapeutic interventions aimed at improving the low levels of LPC, PC, LPA, PG, PI, PGD2, and PGE1 in inflamed colonic tissues may represent a novel potential treatment approach for restoring inflammatory UC and subsequent epithelial healing. Such interventions hold promise for improving clinical practice and may contribute to the development of personalized medicines for a wide range of diseases, including UC.




