Bioinformatics Service for Post-translational Modifications Analysis
Post-translational modifications (PTMs) represent chemical changes to proteins that occur after translation, significantly altering their activity, localization, stability, and interactions. By leveraging advanced bioinformatics and analytical techniques, PTM analysis enables deeper insights into cellular processes, fostering innovations in biology, medicine, and biotechnology.
Submit Your Request Now
×- Overview
- Types of PTM
- Bioinformatics Methrods
- Our Serives
- Advantages
- FAQs
What Is Post-translational Modifications?
Post-translational modification (PTM) refers to the covalent and generally enzymatic modification of proteins during or after protein biosynthesis. PTMs play a key role in many cellular processes. Living cells are constantly exposed to stimuli from their environment and thus have to adapt to rapidly changing conditions. Owing to the mostly static nature of the genome, a highly dynamic and well-regulated system is required to maintain cellular homeostasis. Compared with gene expression and translational control, which are relatively slow (within minutes to hours), PTMs of proteins stand for the major level of regulation, from very fast and reversible processes such as phosphorylation (seconds) to slower and irreversible ones such as certain forms of glycosylation.
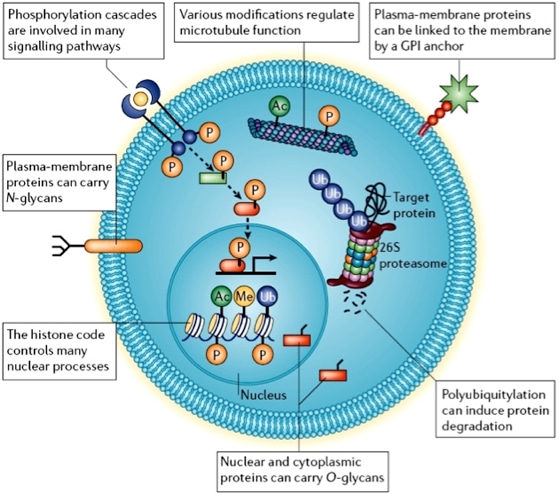 Figure 1. Cellular post-translational modifications(Jensen,2006)
Figure 1. Cellular post-translational modifications(Jensen,2006)
Types of Post-Translational Modifications
Comprehensive List of Common PTMs
- Phosphorylation: Addition of phosphate groups, often regulating enzyme activity.
- Glycosylation: Attachment of sugar moieties, crucial for protein folding and stability.
- Acetylation: Influences gene expression by modifying histones.
- Ubiquitination: Marks proteins for degradation.
- Sumoylation: Modulates protein-protein interactions.
- Methylation: Regulates DNA interactions and chromatin structure.
- Palmitoylation: Anchors proteins to membranes.
See more about other types of PTM in this article "Overview of Post-translational Modification Analysis".
PTM vs Gene Expression
Through PTM of a protein, a multitude of different protein species can be generated, compensating for the incongruity between the relatively low number of encoded genes and the high number of variable regulators required in complex organisms. Currently, more than 450 PTMs are listed in the Uniprot database - with phosphorylation, lysine acetylation, ubiquitination, and proteolytic processing being the most prominent ones. In contrast to gene expression, which merely controls the abundance of proteins, PTMs usually affect their three-dimensional structures, revealing or concealing active sites and interfaces for protein–protein interaction and thus modulating, e.g. subcellular localization, stability, and activity. They act as molecular switches and may initiate and inhibit the interaction of proteins with DNA, cofactors, lipids, as well as other proteins.
PTM involved cellular events
- Gene expression
- Signal transduction
- DNA repair
- Protein-protein interactions
- Protein turnover and localization
- Cellular metabolism
- Cell-cell interactions
- Translocation of proteins across biological membranes
- Communication between internal and external cellular environment
Bioinformatics Analysis Methods of PTMs
PTMs associated database
Through high-resolution MS is a primary source for identifying PTMs, often generating large datasets requiring bioinformatics tools for analysis.Public databases such as UniProt, PhosphoSitePlus, and PTMcode provide curated PTM datasets, including experimental and predicted modification sites.
Table1. Common Databases for PTMs
| Database | Description | Key Features |
|---|---|---|
| PhosphoSitePlus | Comprehensive resource for experimentally verified PTMs, especially phosphorylation. | Disease-specific PTM data, visualization tools |
| UniProt | General protein database including annotations for PTMs. | PTM annotations, cross-referenced datasets |
| PTMcode | Focuses on PTM interactions and cross-talk in proteins. | PTM cross-talk network maps |
| dbPTM | Database for experimentally validated and predicted PTMs. | PTM predictions, literature references |
| Phospho.ELM | Collection of experimentally verified phosphorylation sites. | Functional annotations, kinase-specific data |
| HPRD | Human protein reference database with PTM annotations. | Human-specific PTM data |
| SwissPalm | Database focused on protein palmitoylation. | Palmitoylation-specific PTM sites |
| LysineDB | Database for lysine-specific PTMs like acetylation, ubiquitination, and methylation. | Lysine-centric PTM annotations |
| PLMD | Protein lysine modification database for acetylation and ubiquitination. | Lysine modifications in multiple species |
| GlyGen | Glycosylation database with integrated glycan and protein data. | Glycosylation-specific PTM annotations |
PTMs analysis software
Post-translational modification (PTM) analysis relies on a range of specialized software tools that enhance the identification, prediction, and understanding of PTMs in proteins. Key software includes MaxQuant and Skyline for mass spectrometry (MS)-based PTM quantification, NetPhos, ModPred, and GPS for predicting phosphorylation and other PTM sites using sequence data, and PEAKS Studio for advanced de novo sequencing and novel PTM identification. Databases like PROSITE and Scansite provide motif-based predictions and kinase-substrate relationship insights, while tools like Cytoscape integrate PTM data with protein interaction networks. These tools support applications ranging from high-resolution MS analysis to network-based functional insights, enabling comprehensive PTM research across diverse biological contexts.
Bioinformatics Analysis of PTMs Service
The protein mass spectrometry data analysis services provided by Creative Proteomics include database searching, data quality control, data preprocessing, statistical analysis, functional enrichment analysis, and personalized analysis, among others. The Bioinformatics Analysis of PTM include following services:
Why Choose Creative Proteomics
- Comprehensive and Customized Solutions: Offers end-to-end bioinformatics services tailored for PTM analysis, including data acquisition and detailed interpretation.
- Advanced Technological Capabilities: Utilizes cutting-edge mass spectrometry, machine learning, and robust computational platforms for accurate and efficient analysis.
- Proven Expertise and Versatility: Combines extensive proteomics and bioinformatics expertise with a track record of delivering high-quality, client-specific solutions across academic, clinical, and industrial sectors.
Protein Sequence Analysis FAQs
Why are post-translational modifications important?
Post-translational modifications (PTMs) are essential for regulating protein function, stability, localization, and interactions, playing critical roles in cellular processes such as signal transduction, gene expression, and immune response. PTMs contribute to the functional diversity of the proteome, influencing biological activities and responses to environmental stimuli. Dysregulation of PTMs is often linked to diseases, including cancer, neurodegeneration, and metabolic disorders.
How can bioinformatics help in analyzing post-translational modifications?
Bioinformatics provides computational tools and algorithms to identify, predict, and analyze PTMs. It facilitates the integration of proteomics data with sequence motifs, structural information, and pathway analysis. Bioinformatics enhances PTM research by enabling large-scale analysis of experimental data, predicting modification sites, elucidating functional impacts, and modeling interactions within cellular networks. It also streamlines the interpretation of mass spectrometry results and supports machine learning approaches for advanced predictions.
What tools are available for PTM analysis?
Several tools are commonly used for PTM analysis:
- MaxQuant and Skyline for quantitative mass spectrometry-based PTM identification.
- NetPhos, GPS, and ModPred for sequence-based PTM site prediction.
- PROSITE and Scansite for motif recognition and kinase-substrate interaction analysis.
- PEAKS Studio for de novo sequencing and novel PTM discovery.
- Cytoscape for visualizing PTM-related protein interaction networks.
What are the common challenges in studying post-translational modifications?
- Technical Complexities: Identifying low-abundance or transient modifications is challenging due to limited sensitivity and resolution of analytical methods.
- Data Interpretation: Deciphering the functional roles of PTMs and their interplay within complex biological networks can be difficult.
- Computational Limitations: Predicting PTM sites and analyzing high-dimensional proteomics data require advanced algorithms, large datasets, and significant computational resources.
- Dynamic Nature: PTMs are often context-dependent and can vary with time, cellular conditions, or external stimuli, adding to the complexity of their study.
Learn about other Q&A about other technologies.
Reference
- Jensen ON. Interpreting the protein language using proteomics. Nature Reviews Molecular Cell Biology. 2006 Jun;7(6):391-403.








