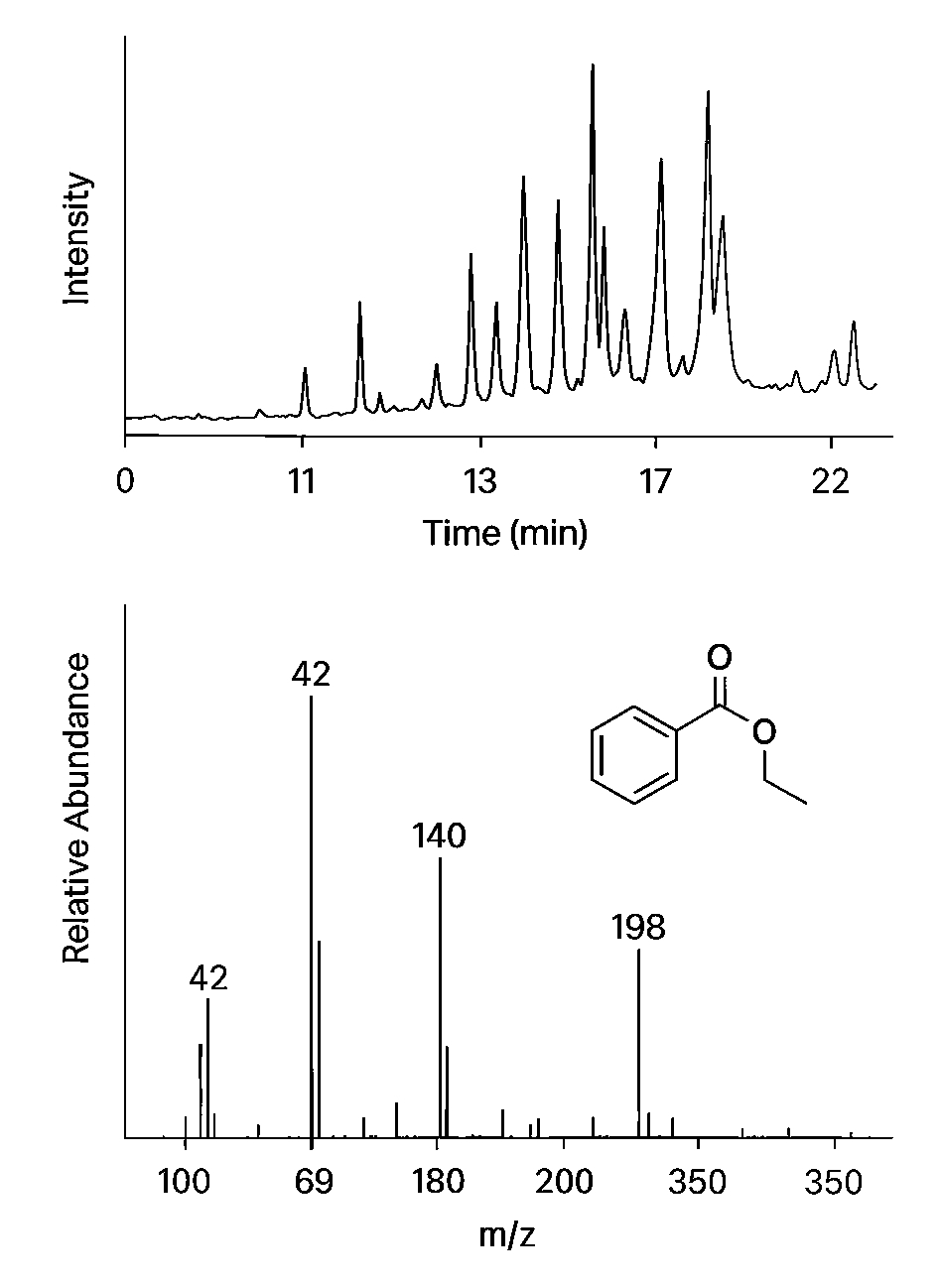
GC-MS/MS Untargeted Metabolomics: Unbiased Metabolite Discovery for Complex Samples
Unlock the full potential of your samples with our advanced GC-MS/MS untargeted metabolomics service. Analyze complex biological, environmental, and food samples for volatile, low-abundance, and novel metabolites that traditional methods miss. Discover deeper insights and biomarkers with confidence.
Our Key Advantages:
- Tailored for volatile & thermally stable metabolites
- Dual mode (Triple Quad & TOF-MS) for broad-spectrum discovery and precise quantification
- Optimized derivatization for expanded metabolome coverage
- High-confidence metabolite identification with curated libraries
- Built-In quality control for consistent, reproducible data
Submit Your Request Now
×
- What We Provide
- Advantages
- Technology Platform
- Sample Requirement
- Demo
- FAQ
Unbiased Metabolite Discovery for Complex Samples
In metabolomics research, many valuable insights are hidden among compounds that are volatile, low-abundance, or not yet cataloged in standard databases. For studies where the metabolic output cannot be predicted in advance—or when known panels are simply not enough—a targeted list limits discovery.
Creative Proteomics provides a GC-MS/MS untargeted metabolomics solution specifically designed for these scenarios. This service focuses on comprehensive detection of thermally stable and volatile metabolites in complex samples, without relying on predefined analyte lists. It's particularly well-suited for studies requiring in-depth metabolic coverage across biological fluids, plant tissues, microbial cultures, food products, or environmental specimens.
Instead of a generic platform, we deliver a methodically tuned workflow that balances analytical depth, reproducibility, and interpretability. With optimized sample derivatization, advanced spectral deconvolution, and high-resolution mass spectrometry, researchers gain access to a more complete picture of metabolic variation—whether the goal is hypothesis generation, biomarker screening, or exploratory systems biology.
What Challenges Does Untargeted Metabolomics Solve?
Metabolomics research often begins with a fundamental uncertainty: which compounds matter, and how can they be reliably captured? For many researchers, the answer isn't clear until the data is in hand—yet standard analytical approaches frequently impose limitations that prevent full metabolic visibility.
- Volatile and Semi-Volatile Metabolites Are Easily Missed
Liquid chromatography–based metabolomics (LC-MS) is widely used, but it is not optimized for thermally stable volatile compounds, which often play key roles in microbial communication, plant defense, or environmental interactions. These molecules tend to escape detection without gas-phase separation and electron ionization—leading to incomplete metabolic coverage.
- Low-Abundance Signals Are Buried in Background Noise
In complex biological or environmental matrices, small but biologically significant metabolites can be masked by high-abundance background compounds. Without the high sensitivity and selectivity of a tandem MS system, these signals are easily lost or misidentified.
- Unknown Compounds Cannot Be Found on a Predefined List
Targeted panels are valuable for quantifying known metabolites—but when the goal is discovery, these panels become a constraint. Predefined analyte lists inherently exclude unexpected or novel metabolites, limiting the potential for pathway exploration and hypothesis generation.
- Reproducibility and Data Quality Remain a Constant Concern
Analytical variation across batches, instruments, or operators can undermine data reliability. Researchers need workflows that not only generate comprehensive metabolite profiles, but also maintain consistency, traceability, and clarity in output.
Choosing the Right Metabolomics Technique
| Feature | GC-MS | GC-MS/MS | LC-MS | HRMS |
|---|---|---|---|---|
| Ideal For | Volatile and semi-volatile metabolites (e.g., alcohols, fatty acids) | Same as GC-MS, but with enhanced quantification for low-abundance metabolites | Polar metabolites (e.g., amino acids, organic acids) | Broad metabolite profiling, including polar and non-polar |
| Sensitivity | High for volatile metabolites | Very high, especially with tandem MS (MS/MS) for low-abundance compounds | High for polar metabolites, less for volatiles | Ultra-high for trace metabolites, with accurate mass |
| Sample Types | Biological fluids, tissues, plant extracts, food | Same as GC-MS, but with better reproducibility for quantification | Blood, plasma, urine, tissues, food, plant samples | Broad, including biological fluids, tissues, environmental samples |
| Metabolite Coverage | Focuses on volatile, small molecules | Same as GC-MS, but also better for quantification of low-abundance compounds | Best for polar, water-soluble metabolites, lipids to an extent | Very high coverage across polar and non-polar metabolites |
| Quantification | Accurate with internal standards | Highly accurate, especially in MRM mode (for specific targets) | Reliable for known metabolites, less for volatiles | Excellent for both known and unknown metabolites |
| Resolution | Moderate, limited by GC column separation | Moderate to high, enhanced with tandem MS (MS/MS) | Moderate to high, enhanced with MS for complex mixtures | Ultra-high resolution for precise metabolite quantification |
| Metabolite Discovery | Limited to volatile and semi-volatile metabolites | High for known and unknown metabolites | High for polar metabolites, but misses volatiles | Best for novel and low-abundance metabolites |
| Technical Complexity | Moderate, requires derivatization for some metabolites | Moderate, but offers more detailed analysis | Moderate, with automated processing options | High, requires specialized expertise |
| Throughput | Moderate, with derivatization steps | Moderate, but can be faster than GC-MS for quantitative work | High, ideal for large sample sets | Low to moderate, based on resolution and complexity |
Why Choose Creative Proteomics?
- Specialized for Volatile & Thermally Stable Compounds
GC-MS/MS is ideal for metabolites that LC-MS often misses—such as alcohols, aldehydes, short-chain fatty acids, and other volatile organics. - Dual Mode: Triple Quad & TOF-MS
Combines targeted-level sensitivity (MRM) with broad-spectrum discovery capability—flexible for both known and unknown metabolites. - Chemically Optimized Sample Derivatization
Custom derivatization (e.g., silylation, methoximation) improves detection of polar or labile metabolites, expanding your metabolome coverage. - High-Confidence Annotation with Curated Libraries
Identification based on NIST, Fiehn, and in-house databases with verified fragmentation and retention index alignment. - Built-In QC and Reproducibility Controls
Internal standards, pooled QCs, and batch effect tracking ensure consistent, trustworthy data—ready for downstream analysis. - Supports a Wide Range of Sample Types
Validated protocols for plasma, serum, urine, tissues, fermentation broths, plant extracts, food products, and more.
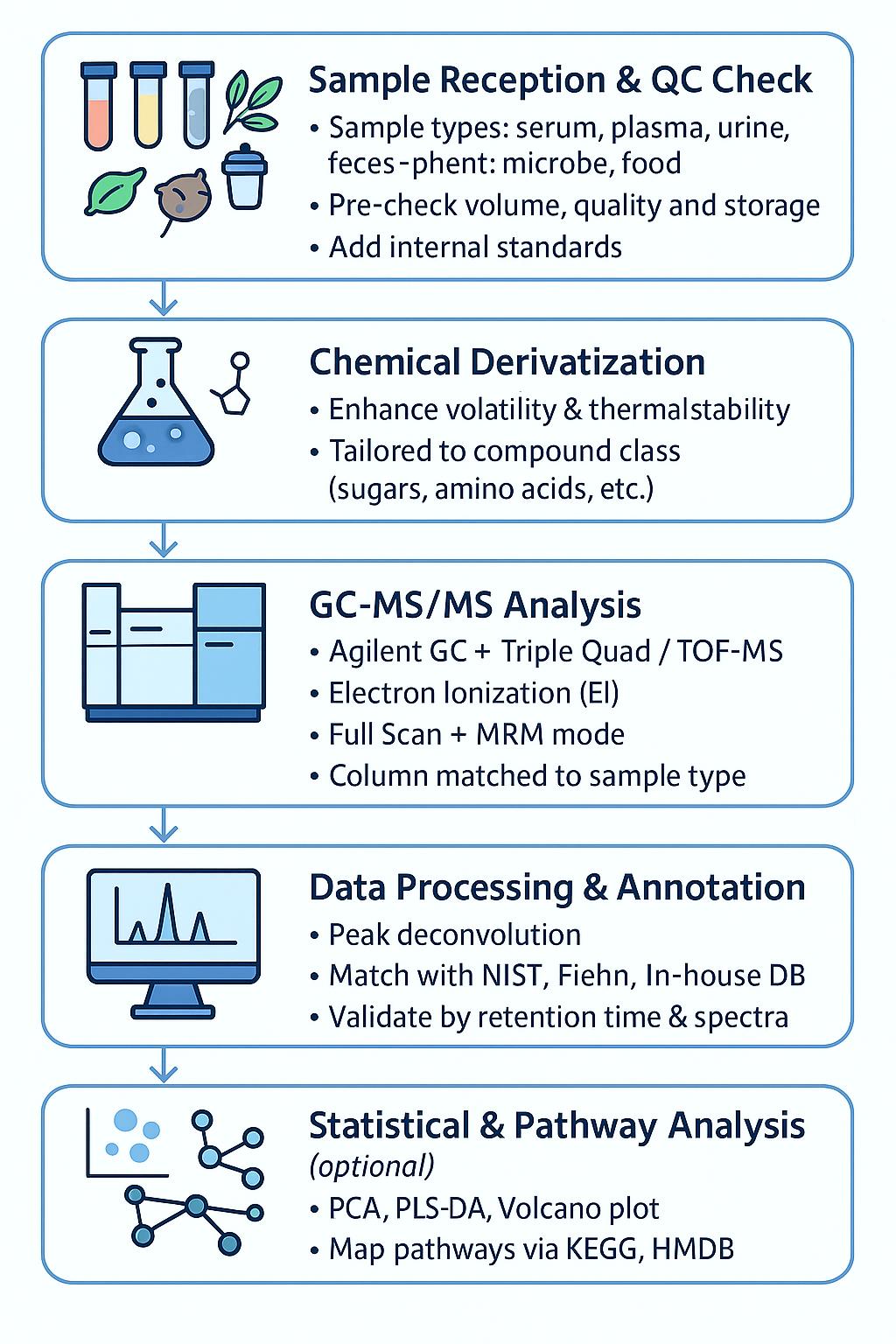
How the Service Works
1. Sample Reception & QC Check
We receive a variety of sample types, including serum, plasma, urine, feces, plant tissue, microbial cultures, and food matrices. Each sample undergoes a quality check to verify volume, quality, and storage conditions. Internal standards are added during preparation.
2. Chemical Derivatization
To enhance volatility and thermal stability, we tailor the derivatization process for specific compound classes such as sugars, amino acids, and organic acids.
3. GC-MS/MS Analysis
The analysis is conducted using an Agilent GC with Triple Quad or TOF-MS instrumentation. We use Electron Ionization (EI) in full scan and MRM modes, with column selection optimized for each sample type.
4. Data Processing & Annotation
The data undergoes peak detection and deconvolution. We match spectra with NIST, Fiehn, and in-house libraries and validate retention time and mass spectral data.
5. Statistical & Pathway Analysis (optional)
For additional insights, we perform statistical analyses such as PCA, PLS-DA, and volcano plots, as well as pathway mapping via KEGG and HMDB.
6. Results Delivery
The final deliverables include an annotated metabolite list with names, formulas, retention times, and intensities, along with a QC report and method summary. Raw and processed data files are available upon request.
Technical Highlights for GC-MS/MS Metabolomics Service
| Feature | Specification |
|---|---|
| Instrumentation | GC-MS/MS (Triple Quad or TOF) |
| Detection Range | C1–C30 compounds, volatiles, semi-volatiles |
| Ionization Mode | EI (Electron Ionization) |
| Coverage | >500 metabolites/sample (typical) |
| Derivatization | MOX, MSTFA, BSTFA (as needed) |
| Output Format | Excel/CSV tables, plots, optional pathway maps |
 7010D GCMS (Figure from Agilent)
7010D GCMS (Figure from Agilent)
Sample Requirements for GC-MS/MS Untargeted Metabolomics Service
| Parameter | Requirement |
|---|---|
| Sample Type | Biological fluids (blood, urine, plasma), tissues, cells, plants, food |
| Volume/Weight | 50–200 µL (liquid) / 10–50 mg (solid) |
| Storage | -80°C (long-term storage) |
| Derivatization | Required for most metabolites (e.g., MSTFA) |
| Min. Metabolite Amount | 1–10 ng per metabolite |
| Preparation | Extraction (methanol:water or chloroform:methanol) and centrifugation |
| Blank Control | Solvent blank (e.g., pure solvent) |
| QC | Pooled QC samples, analyte-spiked controls |
| Internal Standard | Deuterated compounds for quantification |
| Replicates | 3–5 biological, 2–3 technical replicates |
| Processing Time | 2–4 hours (extraction and derivatization) |
Demo Results
FAQ for GC-MS/MS Untargeted Metabolomics Service
How does GC-MS/MS detect low-abundance metabolites in complex samples?
GC-MS/MS achieves high sensitivity for low-abundance metabolites through tandem MS (MS/MS) for targeted detection and internal standards for accurate quantification. Chemical derivatization enhances volatility and thermal stability, ensuring metabolites are detectable even in complex matrices like plasma or tissues.
Why is chemical derivatization important in this analysis?
Derivatization increases volatility and stability of metabolites, especially polar compounds (e.g., amino acids, organic acids). This process ensures better peak resolution and sensitivity, allowing a broader range of metabolites to be analyzed and identified with greater accuracy.
How do mass spectral libraries aid in metabolite identification?
Spectral libraries (e.g., NIST, Fiehn) provide reference fragmentation patterns and retention times to match metabolites against known compounds, ensuring accurate identification. For unknown metabolites, retention index matching and advanced deconvolution help in assigning identities, even for novel compounds.
How do you ensure reproducibility and data quality?
We ensure high data quality through internal standards, pooled QC samples, and regular blank checks to eliminate contamination. Coefficient of variation (CV) and R² values monitor intra- and inter-assay variability, guaranteeing reliable and consistent results.
How does GC-MS/MS handle unknown metabolites?
GC-MS/MS captures both known and novel metabolites by analyzing all compounds in a sample, not just predefined ones. The method's full scan and MRM modes enable detection of unexpected metabolites, and advanced spectral matching helps identify even previously uncharacterized compounds.
How do you validate the biological relevance of identified metabolites?
We validate metabolites through pathway analysis (e.g., KEGG, HMDB) and multivariate statistics (e.g., PCA, PLS-DA) to confirm biological significance. Comparison with literature and experimental replication strengthens the findings, ensuring they reflect real biological phenomena.
How does your service support metabolic pathway exploration?
GC-MS/MS allows for comprehensive detection of both known and novel metabolites, facilitating the exploration of novel pathways. Pathway mapping and multivariate analysis help uncover connections and biological insights, driving hypothesis generation and systems biology research.
How do you analyze metabolites in complex matrices like tissues or food?
We use tailored extraction methods for each matrix type (e.g., methanol:water for tissues, chloroform:methanol for lipids) to maximize recovery. Derivatization protocols ensure volatility, and QC measures maintain data consistency, even with challenging samples like food or plant tissues.
Learn about other Q&A about proteomics technology.
Publications
Here are some of the metabolomics-related papers published by our clients:

- Methyl donor supplementation reduces phospho‐Tau, Fyn and demethylated protein phosphatase 2A levels and mitigates learning and motor deficits in a mouse model of tauopathy. 2023.
- A human iPSC-derived hepatocyte screen identifies compounds that inhibit production of Apolipoprotein B. 2023.
- The activity of the aryl hydrocarbon receptor in T cells tunes the gut microenvironment to sustain autoimmunity and neuroinflammation. 2023.
- Lipid droplet-associated lncRNA LIPTER preserves cardiac lipid metabolism. 2023.
- Inflammation primes the kidney for recovery by activating AZIN1 A-to-I editing. 2023.
- Anti-inflammatory activity of black soldier fly oil associated with modulation of TLR signaling: A metabolomic approach. 2023.
- Plant Growth Promotion, Phytohormone Production and Genomics of the Rhizosphere-Associated Microalga, Micractinium rhizosphaerae sp. 2023.