O-glycosylation of proteins is a common modification with diverse structures and functions. Due to the clinical and biopharmaceutical importance, it is necessary to comprehensively characterize O-glycoproteins including the determination of glycan composition, sequence and attachment site. Site-specific O-glycosylation analysis is key to explore structure-function relationships of glycoproteins, such as in the context of antigenicity and disease progression. Mass spectrometry (MS)-based methods have proven to be the core technique in site-specific O-glycosylation analysis.
Overview of O-Glycosylation Site Occupancy Analysis Service
Because the addition of O-glycans affects intracellular processes including protein folding and maturation, O-glycosylation site occupancy must be measured quantitatively to unravel the macroheterogeneity in glycoprotein populations, which is important for quality control of glycoproteins. Quantitation of O-glycosylation site occupancy can be applied to the fields of diagnostics, glycotherapeutics, and glycolengineering. At Creative Proteomics, we provide a reliable and affordable O-glycosylation site occupancy analysis service for academics, biotechnology, and pharmaceutical communities.
Mass spectrometry (MS) has emerged as a powerful tool for comprehensive analysis of glycoproteins. Creative Proteomics utilizes the most advanced MS platforms and other proteomic techniques to provide the accurate, reliable O-linked glycosylation site occupancy analysis service for identification of truly O-glycosylated peptides.
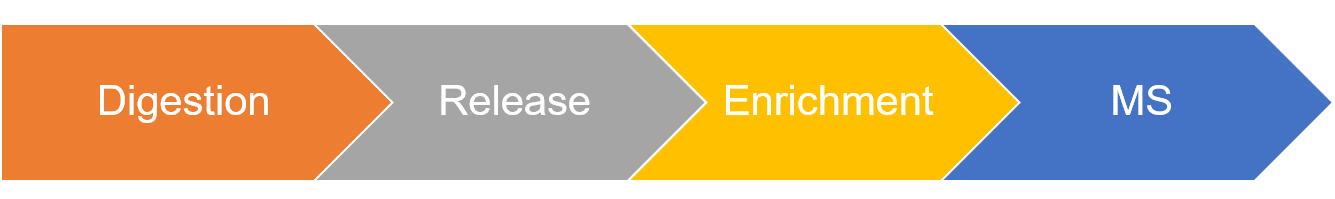
- Protein digestion using enzymes.
- O-Glycopeptide release through β-elimination.
- O-glycopeptide enrichment using HILIC (hydrophilic interaction liquid chromatography).
- Determination of O-linked glycosylation site occupancy using LC-ESI-MS/MS.
Sample Requirement
We work with a range of sample sources as follows.
- Protein: 100 ug
- Cells: 1×107
- Animal tissues: 1 g
- Plant tissues: 200 mg
- Anticoagulated blood (EDTA): 1 mL
- Serum: 0.2-0.5 mL
- Urine: 2 mL
- Microbial sample: 200 mg (dry weight)
Advantages of O-Glycosylation Site Occupancy Analysis
- High-sensitivity, reproducible quantification of rates of O-linked glycan site occupancy
- Work with complex biological matrices, like cells and tissues
- Comprehensive report containing raw data, parameters, data interpretation, and bioinformatics analysis
As one of the leading companies in the proteomics field with years of experience, Creative Proteomics provides glycomics analysis services that can be customized to your needs. Contact us to discuss your project.
Reference
- Holland J W, Deeth H C, Alewood P F. Analysis of O‐glycosylation site occupancy in bovine κ‐casein glycoforms separated by two‐dimensional gel electrophoresis. Proteomics, 2005, 5(4): 990-1002.
- Hoffmann M, Marx K, Reichl U, et al. Site-specific O-glycosylation analysis of human blood plasma proteins. Molecular & Cellular Proteomics, 2016, 15(2): 624-641.


















