Plasma/Serum Proteomics Service
Plasma and serum are the most accessible clinical biofluids for translational research, capturing system-wide protein signals released from multiple tissues and organs. Plasma/serum proteomics enables cohort-scale, quantitative profiling to support biomarker discovery, disease mechanism studies, and treatment-response monitoring. Creative Proteomics provides end-to-end plasma/serum proteomics services (RUO) with standardized workflows and clear deliverables for downstream statistics and validation.
Best-fit uses
- Biomarker discovery & verification in plasma/serum cohorts
- Low-abundance protein profiling with optional deep workflows
- DIA and 4D-DIA quantitative proteomics for reproducible datasets
- Multi-batch study support with QC summaries and consistency checks
- Actionable deliverables: quant matrix + optional pathway modules
Submit Your Request Now
×- Services
- Advantages
- Workflow
- Applications
- Demo
- FAQs
What Is Blood Proteomics
Proteins within the human blood circulation system hold paramount significance, intimately interconnected with all organs, tissues, and cells, their properties serving as direct reflections of the body's pathological and physiological states. A majority of disease-associated biomarkers find their origins intertwined with blood-based proteins. Blood proteomics entails the intricate exploration of protein ensembles residing within the circulating blood, constituting the most intricate facet of the human proteome.
Choice Between Serum and Plasma for Proteomic Analysis
The primary sample types utilized in blood proteomics research are serum and plasma. Some studies employ serum, while others use plasma. This raises the question: serum or plasma? What are the distinctions between these two sample types, and how should we decide which to use?
Serum refers to the translucent yellowish fluid obtained after coagulation of blood, resulting in the separation of fibrinogen and certain coagulation factors from plasma. Plasma constitutes a vital component of blood, presenting as a pale yellow fluid with a complex composition, including substantial amounts of water, proteins, lipids, inorganic salts, among others. Plasma is acquired by centrifugation of blood after the addition of anticoagulants, distinguishing it from serum primarily due to its fibrin content. However, the presence of anticoagulants may potentially influence research outcomes. The decision to employ either serum or plasma for proteomic investigations depends on the research objectives. For instance, when studying a specific coagulation factor, plasma might be the preferred choice.
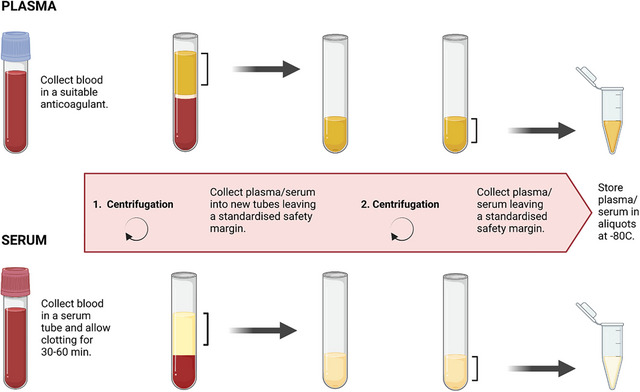 Figure1.Preparation of plasma and serum. (Nieuwland, R et al. 2024)
Figure1.Preparation of plasma and serum. (Nieuwland, R et al. 2024)
Plasma/Serum Proteomics Service at Creative Proteomics
Based on high-abundance protein depletion methods, Creative Proteomics offers various analytical approaches to optimize the detection and analysis of low-abundance proteins. Utilizing mass spectrometry techniques such as Data Independent Acquisition (DIA) and Data-Dependent Acquisition (DDA), we provide high-sensitivity, high-resolution proteomic analysis services for plasma/serum proteins. These approaches effectively remove interfering signals, enhancing the quantification accuracy of low-abundance proteins and enabling researchers to explore biomarkers, disease mechanisms, and other related biological processes in greater depth.
Service Contents
- High Abundant Protein Depletion
- Deep Enrichment and Data Analysis
- Ultimate Deep Proteomics Analysis
- At-a-Glance: Cohort-Ready 4D-DIA Option
High Abundant Protein Depletion
Target application: Basic research and early screening projects with moderate proteomics depth requirements.
Key Features:
- High-Abundance Protein Depletion: Removal of high-abundance proteins (e.g., serum albumin and IgG) from blood samples .
- Protein Identification Depth: Identifies 200-800 proteins, ideal for basic blood proteomics research.
Use Cases:
- Fundamental blood proteomics studies and preliminary biomarker discovery.
- Suitable for small-scale clinical sample analysis or basic research projects.
- Ideal for researchers with limited budgets for initial screening.
Deep Enrichment and Data Analysis
Target application: Mid-sized research institutions or clinical research teams requiring higher proteomic coverage.
Key Features:
- High-Abundance Protein Depletion: Utilizes advanced technologies for optimized high-abundance protein depletion, ensuring enhanced efficiency in removing abundant proteins.
- Low-Abundance Protein Enrichment: Incorporates cutting-edge methods, leveraging superparamagnetic nanoparticle surfaces to improve the detection and enrichment of low-abundance proteins.
- Protein Identification Depth: Identifies 2,000-4,000 proteins, meeting the needs of medium-scale research and clinical studies.
- Machine Learning Analysis: Machine learning algorithms assist in biomarker screening and data analysis, improving the accuracy and efficiency of the research.
Use Cases:
- Medium-scale clinical studies or tumor biomarker screening projects.
- Researchers requiring deep blood proteomics analysis.
- Large population cohort studies, particularly for early disease screening and treatment efficacy evaluation.
Ultimate Deep Proteomics Analysis
Target application: Large-scale clinical research, major biopharmaceutical companies, and projects requiring high proteomic depth and comprehensive data analysis.
Key Features:
- High-Abundance Protein Depletion: Utilizes ultra-sensitive techniques for low-abundance protein enrichment, significantly improving detection sensitivity compared to traditional methods.
- Deep Enrichment Technology: Combines advanced nanoparticle-based methods with high-depth proteomics approaches to further enhance low-abundance protein enrichment, maximizing proteomic coverage.
- Protein Identification Depth: Identifies over 6,000 proteins, suitable for high-depth research and large-scale cohort analysis.
- Advanced Data Analysis: Utilizes fully automated mass spectrometry platforms and machine learning algorithms to comprehensively screen biomarkers, supporting precise analysis of large cohort data.
- No Species Limitations: Suitable for various biological fluid samples, not limited to serum or plasma.
Use Cases:
- Large-scale clinical studies, early tumor screening, and precision medicine projects.
- High-throughput sample analysis, drug development and validation, especially for high-accuracy biomarker screening.
- Suitable for integrating high-depth multi-omics datasets, supporting translational research in the biopharmaceutical industry.
At-a-Glance: Cohort-Ready 4D-DIA Option
For plasma/serum studies where missing values, interference, and batch effects become the main risk, we offer an optional 4D-DIA quantitative proteomics workflow on Bruker timsTOF HT (TIMS-XR) with dia-PASEF®, with CCS-assisted analysis for robust identification and cohort-scale consistency (project-dependent).
Best fit: biomarker discovery/verification, multi-batch cohorts, translational plasma/serum studies.
Designed for 30–200+ sample cohorts (batch-aware QC planning)
Deliverables: Quant matrix + QC report (optional differential/pathway modules)
Our Advantages
- Comprehensive Proteome Coverage: Our service preserves the full informational value of plasma/serum proteome by eliminating the need for high-abundance protein depletion.
- High-Throughput Automation: We employ a fully automated platform capable of processing 96 samples simultaneously, with a streamlined workflow that completes enrichment and peptide digestion in just 7 hours.
- Enhanced Efficiency: Our optimized protocol eliminates the need for peptide fractionation, significantly improving overall process efficiency.
- Rapid Mass Spectrometry Analysis: We deliver fast turnaround times with a throughput of 8-14 minutes per sample in both DDA and DIA modes.
Blood Proteomics Service Workflow
Sample Collection and Preparation
After blood collection, samples undergo preprocessing steps, including centrifugation, quality screening, protein extraction, and dehydration, ensuring sample purity and stability. Based on experimental needs, appropriate methods for high-abundance protein depletion are selected to optimize the analysis.
Mass Spectrometry Analysis
Following preprocessing, samples are introduced into a mass spectrometer for analysis. Mass spectrometry analysis is the cornerstone technology of clinical blood proteomics, primarily employing ionization, separation, and detection to analyze protein components within the sample. Commonly used mass spectrometer is ESI-MS. The analysis yields an initial raw mass spectrum.
Data Analysis MS raw data will be analyzed by certain software. With proper protein database search, we will get protein identification and relative abundance results. Bioinformatics AnalysisTo derive statistically meaningful results from proteomics datasets, further data analysis is conducted. This encompasses differential analysis, clustering analysis, pathway analysis, and more.
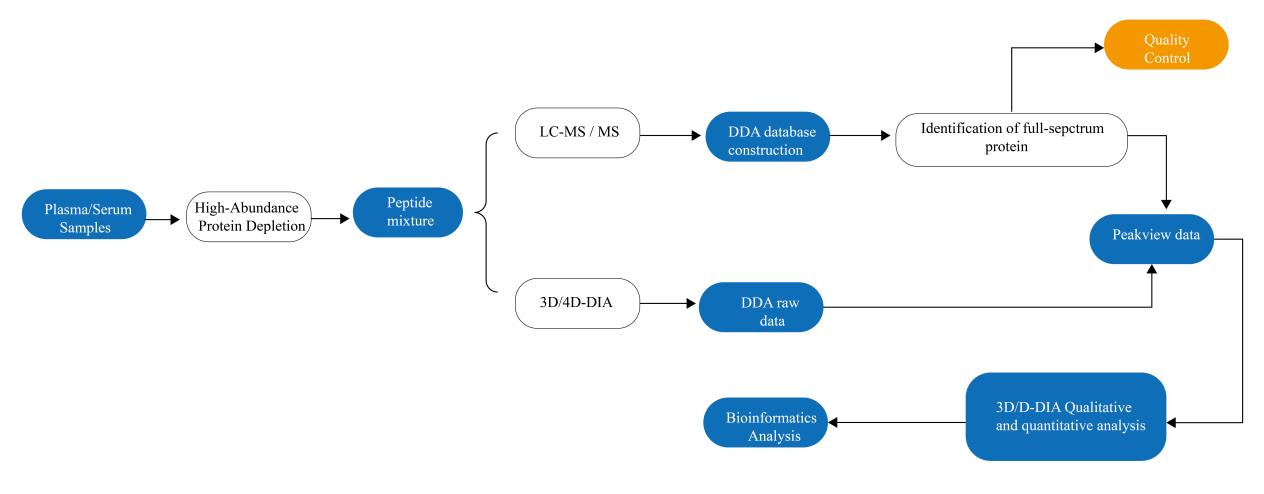
Result Output
The output of blood proteomics includes information at two levels. First, a quantitative results table is generated, listing detected proteins in the samples and their relative abundances. We can also provide MS raw files if request. Second, a comprehensive bioinformatics analysis for further interpretation of obtained protein identified and quantified data. Experimental steps
- Relevant experiment parameters
- Raw data
- Proteomics analysis results
Technical Platforms

Thermo Q ExactiveTM series

AB Sciex 6500+

Thermo Orbitrap Fusion Lumos

Bruker timsTOF Pro
Sample Requirements
Whether in plasma proteomics or serum proteomics, the process begins with preparing plasma or serum from blood. Plasma is obtained by centrifuging blood with anticoagulants added, while serum is obtained by centrifuging coagulated blood. Thus, researchers need to choose the appropriate blood processing method based on their research objectives. Blood samples should be stored at -80°C and should not undergo repeated freeze-thaw cycles.
| Sample Type | Plasma/Serum |
|---|---|
| Quantify | > 100 μL |
Appliacations of Blood Proteomics
Disease Diagnosis and Early Detection
Blood proteomics analyzes protein changes in plasma and serum to identify disease-specific biomarkers, providing critical insights for early disease diagnosis.
Cancer Biomarker Discovery
Mass spectrometry-based analysis of blood proteomes enables the screening of cancer-related biomarkers, supporting early cancer detection, diagnosis, and treatment monitoring.
AI-Driven Proteomics Analysis
Integrating blood proteomics with artificial intelligence (AI), leveraging machine learning and deep learning techniques, enhances the analysis of large-scale proteomic data, improves biomarker identification accuracy, and advances personalized medicine research.
Exosome Proteomics Research
Analyzing the protein composition of exosomes in blood explores their potential for early cancer diagnosis, offering novel biomarker sources for disease screening.
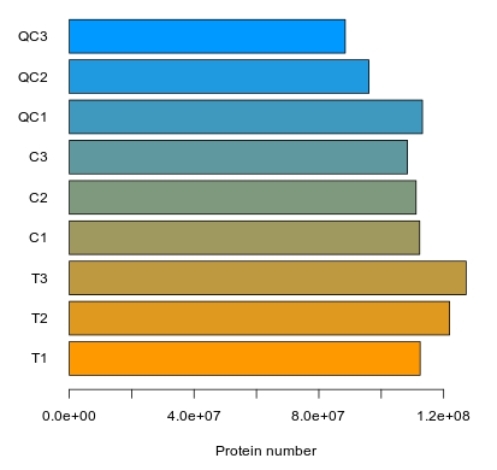
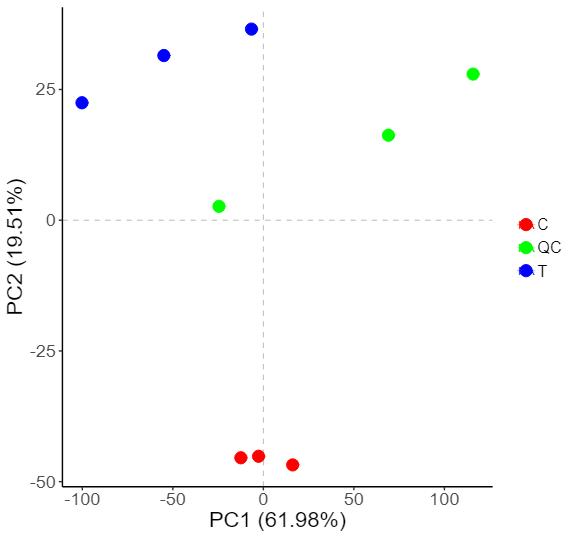
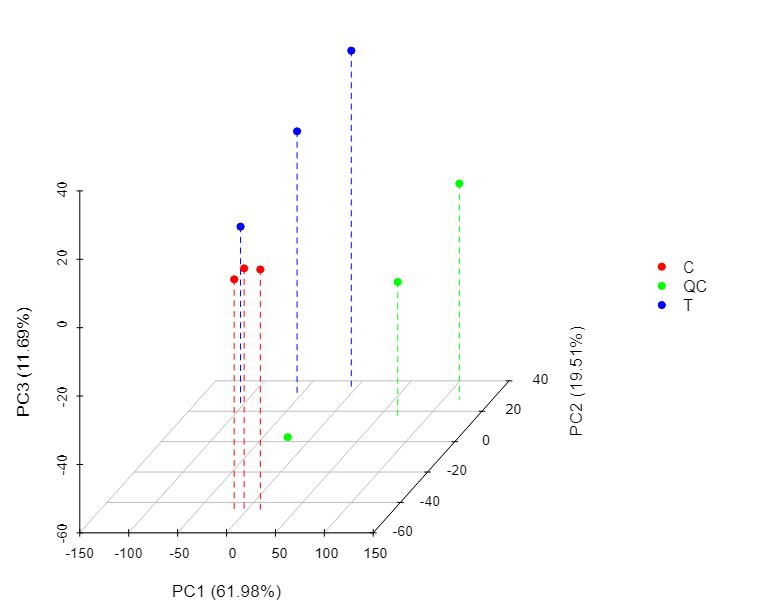
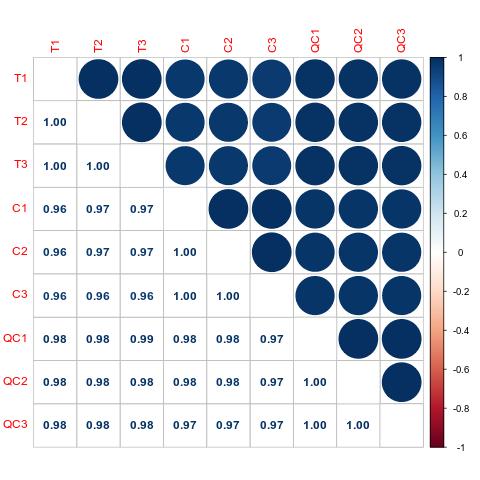
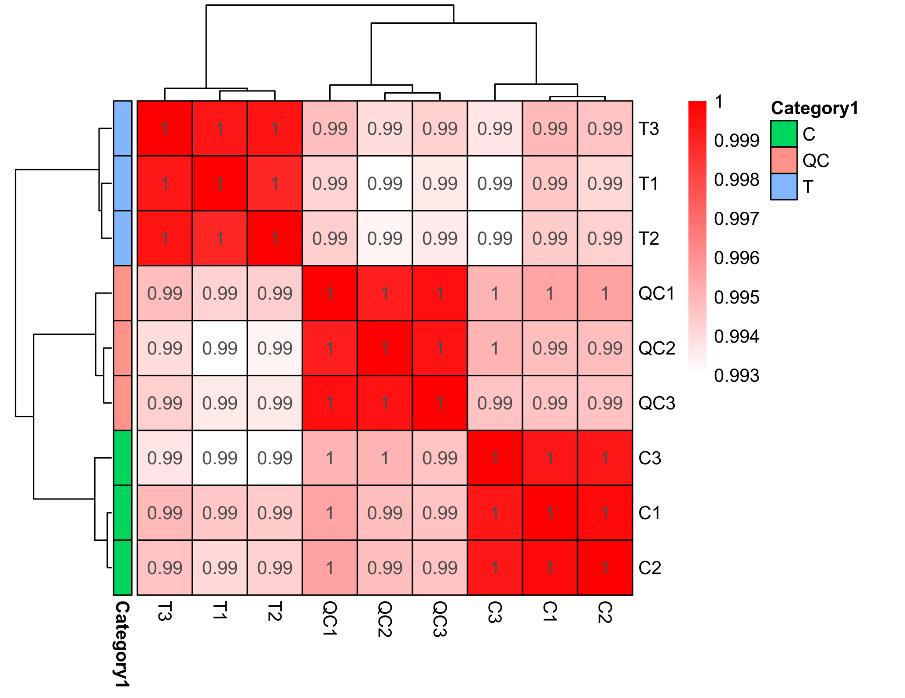
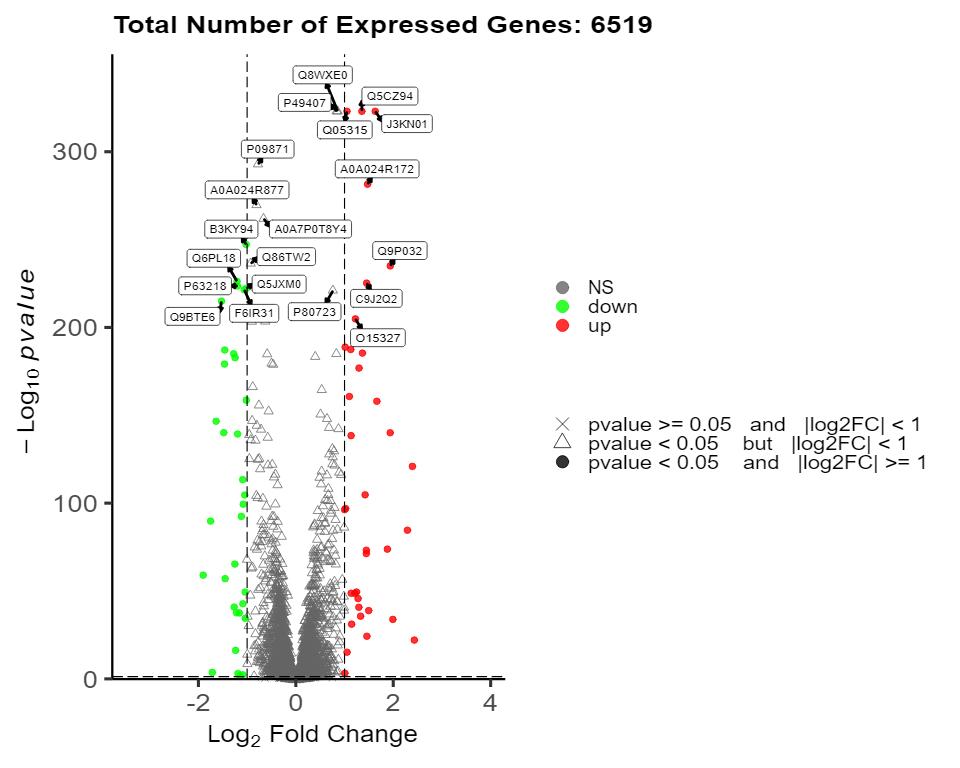
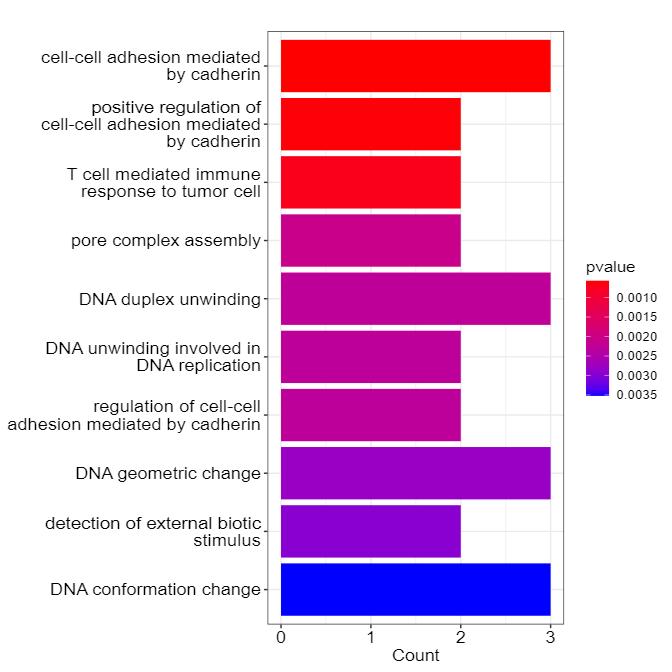
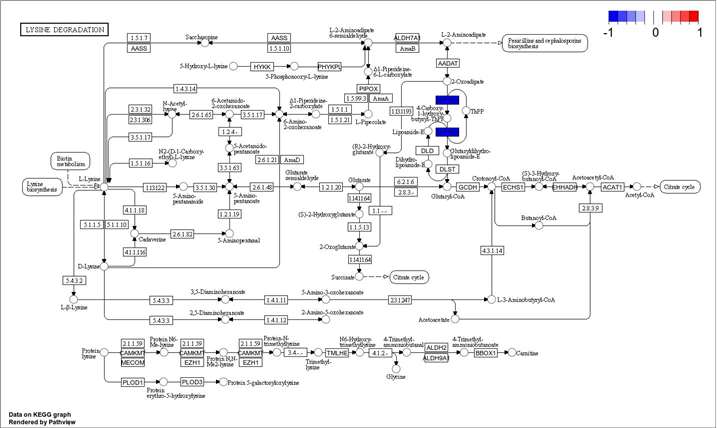
Demo Results
Blood Proteomics FAQs
What is the significance of serum and plasma proteomics in disease research?
Serum and plasma proteomics are critical subfields of proteomic research that focus on analyzing the complete protein expression profiles in plasma and serum. By identifying differential proteins and disease-associated proteins, this approach opens new pathways for understanding the pathophysiological mechanisms of major diseases, discovering early diagnostic biomarkers, and uncovering potential drug targets. During disease progression, low-abundance proteins secreted into plasma and serum play a key role in early diagnostics and pathogenesis.
How is data analyzed in plasma/serum proteomics?
Data is analyzed using bioinformatics tools for protein identification, quantification, and functional annotation. Statistical analysis is performed to identify significant changes in protein expression.
Can plasma/serum proteomics identify biomarkers?
Yes, it is widely used for biomarker discovery by comparing protein profiles between healthy and diseased samples. Identified biomarkers can be validated in larger cohorts.
What is 4D-DIA proteomics and when is it preferred for plasma/serum cohorts?
4D-DIA proteomics combines TIMS ion mobility (CCS) with DIA to add an extra separation dimension before fragmentation, supporting more consistent quantification in complex matrices (project-dependent). It is often preferred for plasma/serum cohorts (typically 30–200+ samples) where missing values, interference, and multi-batch stability are key risks.
How does TIMS/CCS help reduce interference in plasma proteomics?
TIMS separates ions by mobility before MS/MS, and CCS provides an additional identifier that can support peptide confidence and cross-run alignment (project-dependent). In plasma/serum, this extra dimension can help reduce co-fragmentation interference and improve cohort-scale consistency compared with 3D workflows.
When should I consider CPDB3000™ for low-abundance biomarker studies?
Consider CPDB3000™ when your study depends on mid-to-low abundance plasma/serum proteins and you want a workflow designed to reduce high-abundance masking via low-abundance enrichment (project-dependent) + 4D-DIA (TIMS/CCS + DIA). It is commonly used for biomarker discovery/verification and large-cohort translational studies where data completeness and batch-aware QC matter.
Learn about other Q&A about other technologies.
Publications
Here are some publications in Proteomics research from our clients:

- Microglia-mediated synaptic pruning in the nucleus accumbens during adolescence: A preliminary study of the proteomic consequences and putative female-specific pruning target. 2023. https://doi.org/10.1101/2023.05.03.539317
- Regulation of toxic shock syndrome toxin‐1 by the accessory gene regulator in Staphylococcus aureus is mediated by the repressor of toxins. 2019. https://doi.org/10.1111/mmi.14353
- The yeast protein Mam33 functions in the assembly of the mitochondrial ribosome. 2019. https://doi.org/10.1074/jbc.RA119.008476
- Identification of a novel anti-HIV-1 protein from Momordica balsamina leaf extract. 2022. https://doi.org/10.3390/ijerph192215227
- Pressure overload induces ISG15 to facilitate adverse ventricular remodeling and promote heart failure. 2023. https://doi.org/10.1172/JCI161453
References
- Nieuwland, R., & Siljander, P. R. (2024). A beginner's guide to study extracellular vesicles in human blood plasma and serum. Journal of extracellular vesicles, 13(1), e12400. https://doi.org/10.1002/jev2.12400
- Tao, Q. Q et al. (2024). Alzheimer's disease early diagnostic and staging biomarkers revealed by large-scale cerebrospinal fluid and serum proteomic profiling. Innovation (Cambridge (Mass.)), 5(1), 100544. https://doi.org/10.1016/j.xinn.2023.100544
- Oh, H. S et al. (2023). Organ aging signatures in the plasma proteome track health and disease. Nature, 624(7990), 164–172. https://doi.org/10.1038/s41586-023-06802-1