Coenzyme I Analysis Service
Coenzyme I dynamics are central to cellular energy, redox balance, and metabolic adaptation. Creative Proteomics offers specialized assays to precisely quantify NAD⁺/NADH and related metabolites, empowering researchers to decode complex biological systems and optimize bioprocesses.
Our Advantages:
- Ultra-sensitive detection down to 0.01 ng/mL for NAD⁺/NADH and related compounds
- Comprehensive profiling covering NAD⁺, NADH, NADP⁺, NADPH, NMN, NR, and more
- Robust analysis across plasma, tissues, cells, and fermentation matrices
- Customizable methods to fit diverse experimental needs
- Expert data interpretation and pathway insights for actionable results
Submit Your Request Now
×
Deliverables You Can Expect:
- High-sensitivity quantification of NAD⁺, NADH, NADP⁺, NADPH, NMN, NR, and more.
- Detailed ratio analysis (e.g. NAD⁺/NADH) for metabolic insights.
- Comprehensive data reports with raw files and interpretation.
- Optional pathway enrichment analysis for biological context.
- Guidance on experimental design and result application.
- What We Provide
- Advantages
- Technology Platform
- Sample Requirement
- Demo
- Case
- FAQ
What are Coenzyme I Compounds?
Coenzyme I compounds refer primarily to a group of nicotinamide-based redox cofactors, including NAD⁺ (nicotinamide adenine dinucleotide) and its phosphate form NADP⁺, along with their reduced counterparts NADH and NADPH. These cofactors are essential molecular agents in redox reactions, acting as electron carriers in a wide range of metabolic and biosynthetic pathways.
Biochemically, Coenzyme I compounds serve critical roles in:
- Energy production via glycolysis, the tricarboxylic acid (TCA) cycle, and oxidative phosphorylation
- Biosynthesis of lipids, nucleotides, and amino acids
- Cellular antioxidant defense through NADPH-driven regeneration of reduced glutathione
- Gene expression regulation and epigenetics via NAD⁺-dependent enzymes (e.g., sirtuins and PARPs)
Due to their central involvement in fundamental cellular processes, accurate quantification of Coenzyme I metabolites provides valuable insights into metabolic states, redox balance, and biochemical responses to environmental or experimental stimuli.
These molecules are not stored long-term but are dynamically interconverted, making their real-time analysis essential for interpreting physiological and biochemical phenomena across biological systems.
Why Analyze Coenzyme I?
Understanding the balance and dynamics of Coenzyme I and its associated metabolites is critical for studies involving:
- Cellular energy metabolism: Evaluate ATP production pathways through NAD⁺/NADH ratios.
- Oxidative stress research: Quantify NADPH levels in relation to reactive oxygen species (ROS) defense.
- Metabolic flux studies: Assess how perturbations in coenzyme levels affect central carbon metabolism.
- Drug metabolism & pharmacodynamics: Examine coenzyme fluctuations following compound treatment or environmental stress.
Coenzyme I Analysis Service Offered by Creative Proteomics
Creative Proteomics provides targeted metabolomics services for coenzyme I analysis using advanced HPLC and MS technologies. Our services support researchers in biochemistry, molecular biology, pharmacology, and related fields with precise and reliable data.
We quantify both oxidized (NAD⁺) and reduced (NADH) forms of nicotinamide adenine dinucleotide to study NAD⁺ metabolism, its cellular roles, and disease associations. Using HPLC and MS, we deliver accurate, reproducible measurements in various biological samples.
Essential for mitochondrial function and antioxidant defense, CoQ10 is analyzed using HPLC and UV-Vis Spectroscopy. We measure CoQ10 levels in blood, tissue, and cell lysates, supporting research on energy metabolism, oxidative stress, and cardiovascular health.
As a precursor to NAD⁺, niacin levels are measured using HPLC with fluorescence detection. Our analysis helps investigate vitamin B3 status and its role in metabolic health and NAD⁺ biosynthesis.
We provide sensitive quantification of nicotinamide using HPLC with fluorescence or MS detection to support research into NAD⁺ metabolism, stress response, and aging.
Custom Analytical Solutions
Tailored services are available to meet specific research needs, including specialized protocols and unique sample types. Our expert team can design and implement custom workflows to support your project goals.
List of Detected Coenzyme I and Related Metabolites
| Compound | Biological Role | Primary Metabolic Pathways |
|---|---|---|
| NAD⁺ (Nicotinamide Adenine Dinucleotide) | Oxidized electron acceptor in redox reactions | Glycolysis, Tricarboxylic Acid (TCA) Cycle, Oxidative Phosphorylation |
| NADH | Reduced electron donor, drives ATP synthesis | Electron Transport Chain (ETC), Mitochondrial Respiration |
| NADP⁺ (Nicotinamide Adenine Dinucleotide Phosphate) | Oxidized coenzyme for biosynthetic reactions | Pentose Phosphate Pathway, Fatty Acid & Nucleotide Biosynthesis |
| NADPH | Reduced coenzyme providing electrons for biosynthesis and antioxidant defense | Fatty Acid Synthesis, Glutathione Reduction, Detoxification Pathways |
| Niacin (Vitamin B3) | Dietary precursor for NAD⁺ biosynthesis | De Novo NAD⁺ Biosynthesis |
| Nicotinamide | Salvage pathway intermediate for NAD⁺ synthesis | NAD⁺ Salvage Pathway |
| Nicotinamide Riboside | Alternative NAD⁺ precursor | Nicotinamide Riboside Kinase (NRK) Pathway |
| Nicotinamide Mononucleotide (NMN) | Direct NAD⁺ precursor | NAD⁺ Biosynthesis Intermediate |
| Nicotinic Acid Adenine Dinucleotide (NAAD) | Intermediate in NAD⁺ biosynthesis | Preiss-Handler NAD⁺ Salvage Pathway |
| Coenzyme Q10 (Ubiquinone) | Lipid-soluble electron carrier in mitochondria | Mitochondrial Respiratory Chain (Complex I, II, III) |
Advantages of Coenzyme I Assay
- Ultra-low Detection Limits: Sensitive quantification down to 0.1 ng/mL, enabling detection of trace metabolites in complex samples.
- Wide Dynamic Range: Measures concentrations across 4+ orders of magnitude for versatile sample compatibility.
- High Precision: Consistent results with coefficients of variation below 5%, ensuring reproducibility across analyses.
- Extensive Metabolite Coverage: Simultaneous quantification of over 10 key Coenzyme I metabolites and intermediates.
- Minimal Sample Volume: Requires only small sample amounts, preserving precious biological material.
- Broad Matrix Compatibility: Accurate analysis across plasma, tissue, cell lysates, and other biological matrices.
Methods and Instrumentation for Coenzyme I Analysis
Agilent 6495C Triple Quadrupole LC-MS/MS
- Ultra-high sensitivity for low-abundance NAD⁺ species
- Dynamic MRM capability for simultaneous multi-analyte detection
- LOD ≤ 0.01 ng/mL for NAD⁺/NADH
- Wide linear range (0.01 – 1,000 ng/mL)
 Agilent 6495C Triple quadrupole (Figure from Agilent)
Agilent 6495C Triple quadrupole (Figure from Agilent)
Agilent 1260 Infinity II HPLC
- Excellent separation of NAD⁺/NADH without derivatization
- UV detection at 260 nm or fluorescence detection for improved sensitivity
- Retention time RSD ≤ 0.3% for stable reproducibility
 Agilent 1260 Infinity II HPLC (Figure from Agilent)
Agilent 1260 Infinity II HPLC (Figure from Agilent)
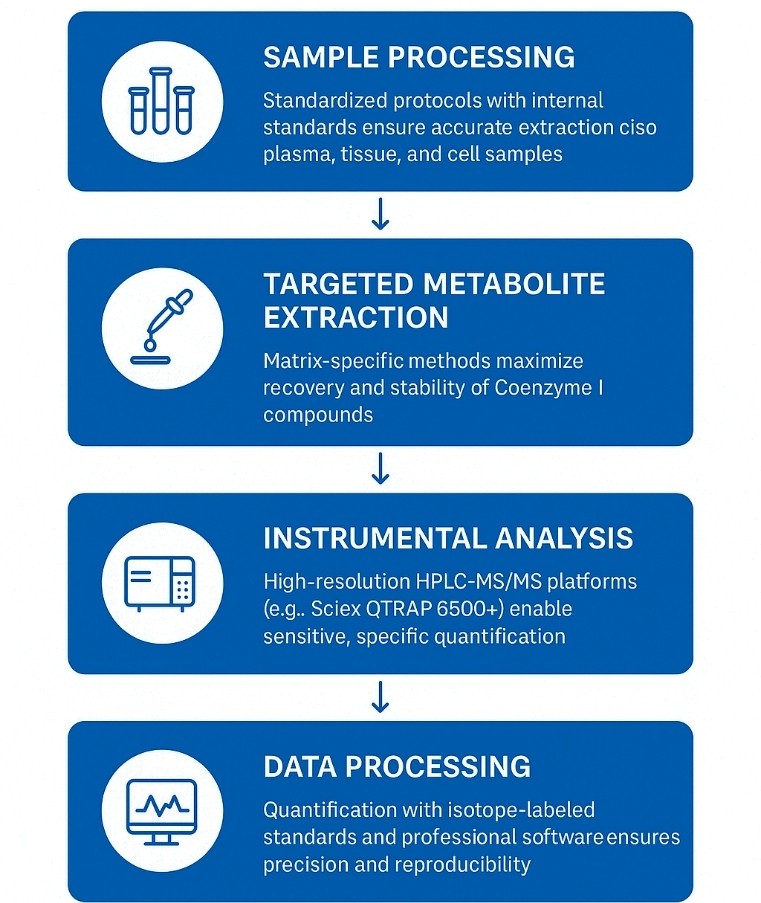
Our Coenzyme I Assay Workflow
1. Sample Preparation & Quality Control
Biological samples (e.g., plasma, tissue, cells) are processed under cold-chain conditions with internal standards to correct for matrix effects. Each batch includes blanks, QCs, and calibration samples for data reliability.
2. Targeted Metabolite Extraction
Matrix-specific protocols minimize degradation and maximize recovery of Coenzyme I compounds, ensuring consistent extraction across sample types.
3. Instrumental Analysis
Samples are analyzed via HPLC-MS/MS. Key parameters like polarity, collision energy, and retention time are optimized per analyte for high sensitivity and specificity.
4. Data Processing & Quantification
Raw data are processed using Analyst or MultiQuant with isotope-labeled standards. Peak integration and baseline correction are manually reviewed for accuracy.
5. Statistical Analysis & Interpretation
Results are assessed for linearity, recovery, and reproducibility. Optional statistical and pathway analysis supports deeper biological interpretation.
Sample Requirements for Coenzyme I Analysis Service
| Sample Type | Minimum Volume / Weight | Recommended Handling & Notes |
|---|---|---|
| Plasma / Serum | ≥ 50 µL | Use EDTA anticoagulant. Freeze immediately at -80°C. |
| Cell Culture Pellet | ≥ 1 × 10⁶ cells | Harvest, wash with cold PBS, and freeze at -80°C. |
| Tissue (Animal or Plant) | ≥ 10 mg | Flash-freeze in liquid nitrogen. Store at -80°C. |
| Fermentation Broth | ≥ 200 µL | Filter if necessary. Store at -80°C. |
| Urine | ≥ 200 µL | Collect in sterile container. Freeze at -80°C. |
| Bacterial Pellet | ≥ 1 × 10⁸ cells | Wash and freeze immediately at -80°C. |
Demo Results
Coenzyme I Analysis Service Case Study

Title: Comparative metabolite profiling of salt sensitive Oryza sativa and the halophytic wild rice Oryza coarctata under salt stress.
Journal: Plant‐Environment Interactions
Published: 2024
- Study Summary
- Background
- Methods
- Results
- Reference
This study explores how the halophytic wild rice O. coarctata maintains metabolic stability under salt stress compared to the salt-sensitive cultivated rice O. sativa. The findings highlight significant differences in nicotinate and nicotinamide metabolism, which are directly linked to Coenzyme I (NAD⁺/NADH) dynamics and cellular stress responses.
Soil salinity severely limits rice productivity worldwide. While O. sativa suffers significant yield loss under saline conditions, O. coarctata thrives in high-salt environments. Understanding metabolic adaptations, especially involving NAD⁺/NADH balance, is crucial for developing salt-tolerant crops.
Researchers conducted untargeted metabolomics using LC–MS to profile root metabolites from both rice species under control and 120 mM NaCl stress. Particular emphasis was placed on changes in nicotinate and nicotinamide metabolism, pathways closely tied to Coenzyme I (NAD⁺/NADH) levels and cellular redox homeostasis.
![]() Creative Proteomics offers targeted and untargeted Coenzyme I analysis services to support similar research needs. Our capabilities include:
Creative Proteomics offers targeted and untargeted Coenzyme I analysis services to support similar research needs. Our capabilities include:
- Ultra-sensitive quantification of NAD⁺, NADH, nicotinamide, nicotinate, NMN, NR, and related metabolites
- High-resolution LC-MS/MS platforms such as Agilent 6495C Triple Quadrupole, achieving LOD as low as 0.01 ng/mL
- Comprehensive pathway enrichment analysis to link Coenzyme I fluctuations to metabolic stress responses
- Customized method development for complex matrices like plant tissues
Metabolic Profiles Comparison:
Metabolite Identification: LC-MS analysis identified 1012 metabolites in the roots of Oryza coarctata and Oryza sativa under control and salt stress conditions.
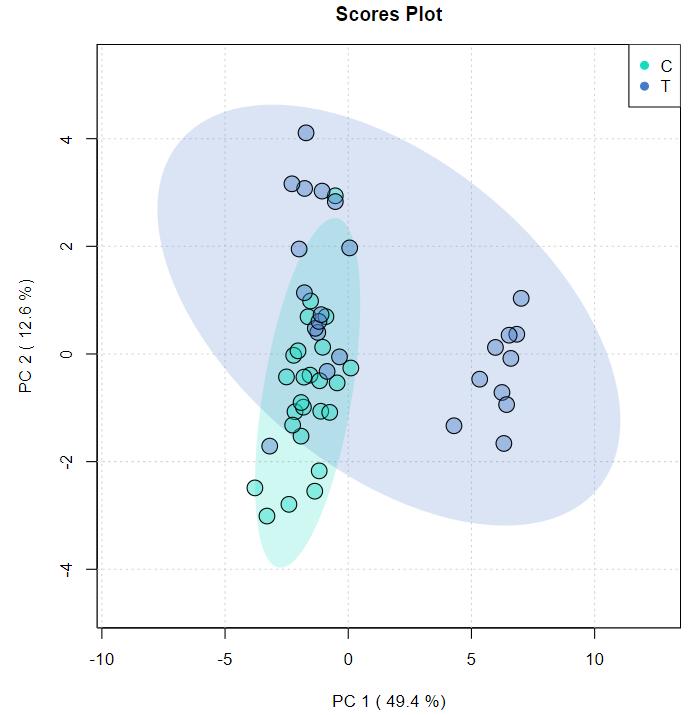
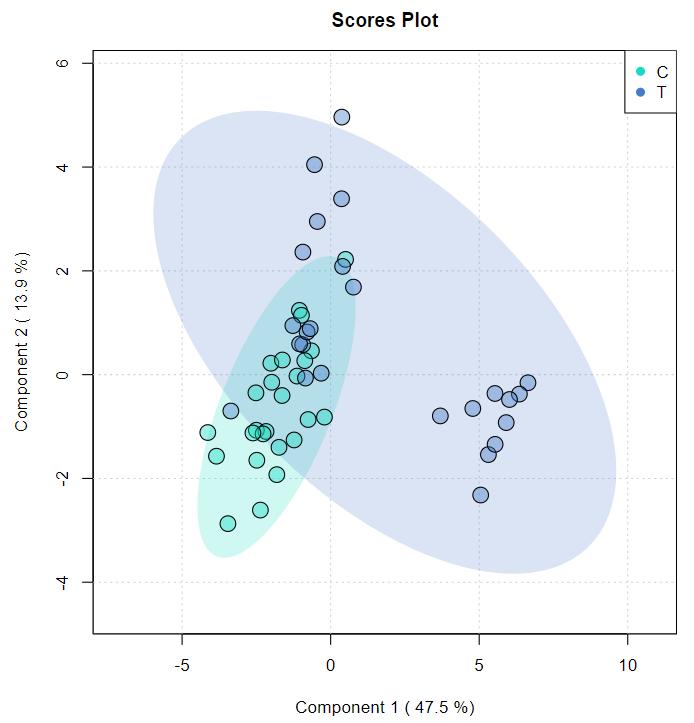
Principal Component Analysis (PCA) and Hierarchical Clustering: Both PCA and hierarchical clustering revealed distinct metabolite clusters between control and salt-stressed samples in both species.
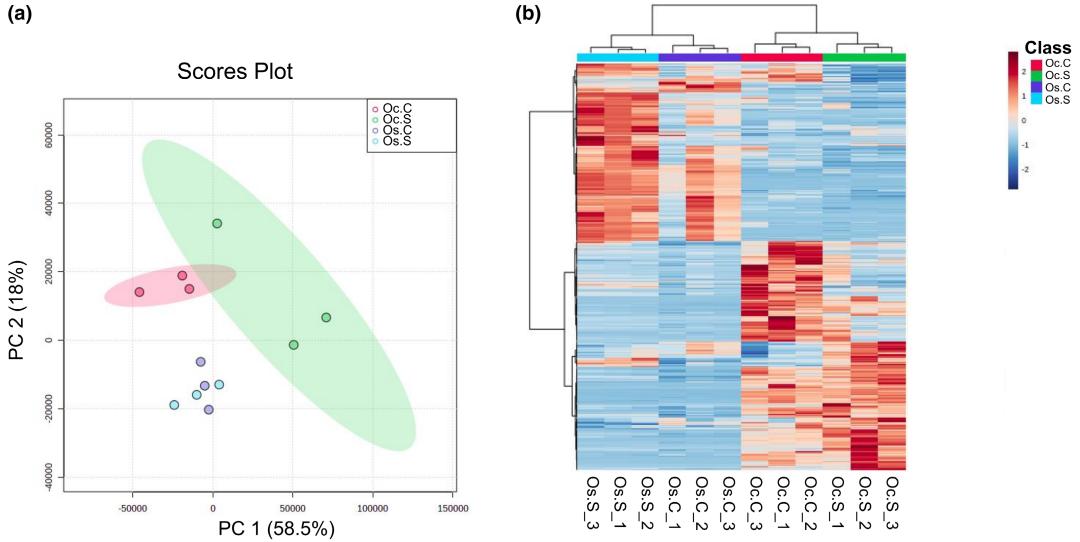 (a) Score plot from PCA analysis of metabolite profiles of Oryza sativa and Oryza coarctata without and with salt stress samples. (b) Hierarchical clustering for the top 500 metabolites of Oryza sativa and Oryza coarctata without and with salt stress samples.
(a) Score plot from PCA analysis of metabolite profiles of Oryza sativa and Oryza coarctata without and with salt stress samples. (b) Hierarchical clustering for the top 500 metabolites of Oryza sativa and Oryza coarctata without and with salt stress samples.
Effects of Salt Stress:
General Findings: Under control conditions, O. coarctata had 380 differentially accumulated metabolites compared to O. sativa. Salt stress increased this number to 436, with 190 metabolites common across conditions. O. coarctata showed downregulation of most metabolites in response to salt, while O. sativa exhibited upregulation.
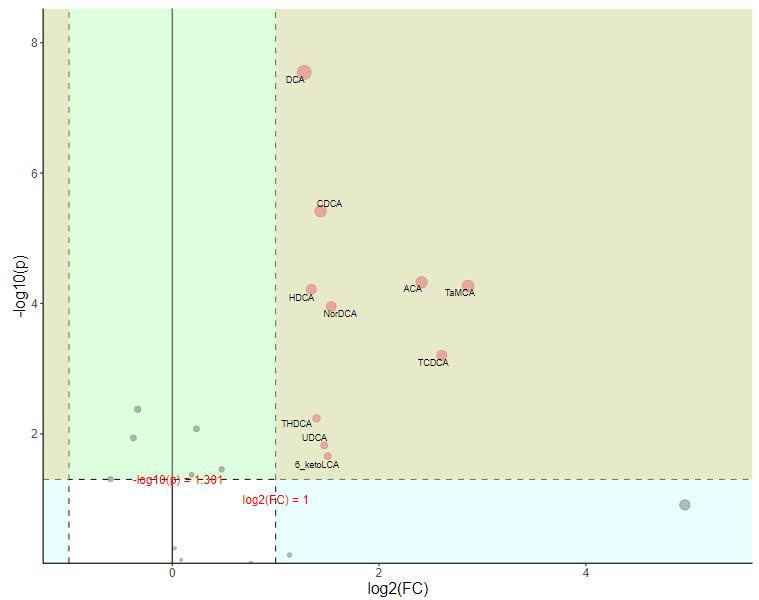
Differential Metabolite Analysis:
- Oc.C vs Os.C (Control Conditions): Higher levels of itaconate, vanillic acid, and xanthin compounds in O. coarctata compared to O. sativa.
- Oc.S vs Oc.C (Salt Stress in O. coarctata): Salt stress led to downregulation of glycerolipids and phospholipids, while amino acids like leucine, phenylalanine, and tyrosine were upregulated.
- Os.S vs Os.C (Salt Stress in O. sativa): Slight upregulation of xanthin compounds and some amino acids under salt stress. Glycerolipids and phospholipids showed minimal changes.
- Oc.S vs Os.S (Salt Stress Comparison): Itaconate, phosphatidylglycerol, and tyrosine were upregulated in O. coarctata compared to O. sativa. Notable differences in the regulation of various phospholipids were observed.
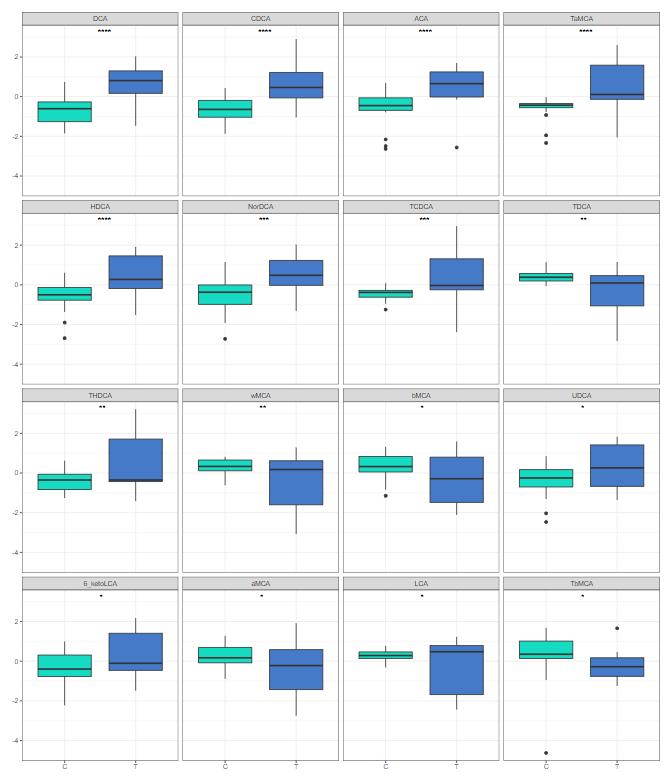
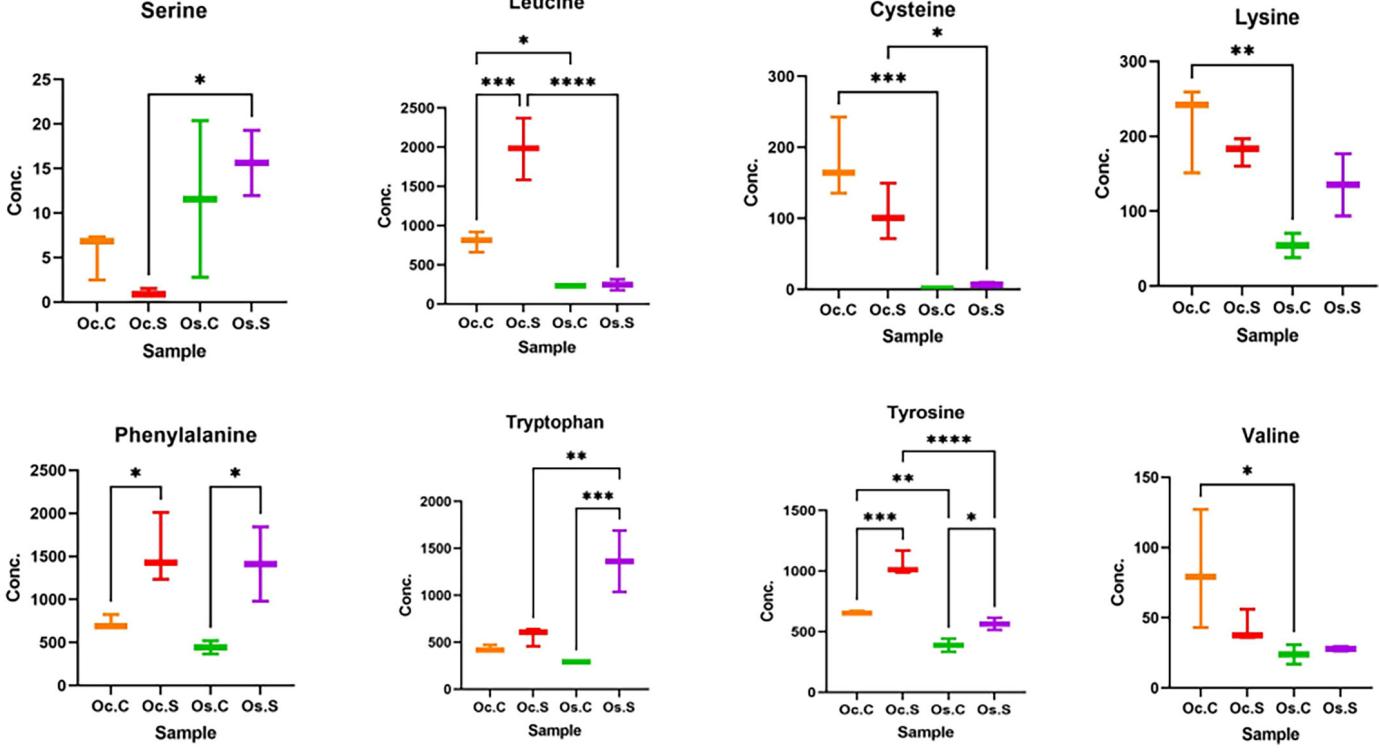 Comparison of amino acid levels in four sample groups: Oc.C, Oc.S, Os.C, and Os.S. Statistical analysis was performed using one-way ANOVA followed by Šídák multiple comparisons test. p < .05 were considered significant. * denotes .01 < p < .05, ** denotes.001 < p < .01, *** denotes .0001 < p < .001, **** denotes p < .0001.
Comparison of amino acid levels in four sample groups: Oc.C, Oc.S, Os.C, and Os.S. Statistical analysis was performed using one-way ANOVA followed by Šídák multiple comparisons test. p < .05 were considered significant. * denotes .01 < p < .05, ** denotes.001 < p < .01, *** denotes .0001 < p < .001, **** denotes p < .0001.
Pathway Enrichment Analysis:
Activated Pathways: Amino acid, fatty acid, and carbohydrate metabolism pathways were significantly activated in both genotypes under salt stress.
O. coarctata: Enhanced sphingolipid metabolism, phenylpropanoid biosynthesis, and eicosanoid accumulation. Higher levels of secondary metabolites like phenylpropanoids and eicosanoids.
O. sativa: Altered biosynthesis of unsaturated fatty acids, nicotinate, nicotinamide, and beta-alanine metabolism. Increased levels of some TCA cycle intermediates and decreased activity in the pentose phosphate pathway.
Reference
- Tamanna, Nishat, et al. "Comparative metabolite profiling of salt sensitive Oryza sativa and the halophytic wild rice Oryza coarctata under salt stress." Plant‐Environment Interactions 5.3 (2024): e10155. https://doi.org/10.1002/pei3.10155
FAQ of Coenzyme I Analysis Service
Can Coenzyme I levels vary significantly between different tissue types?
Yes. Coenzyme I levels can differ greatly between tissues due to variations in metabolic activity, mitochondrial density, and redox demands. For instance, liver and muscle often have higher NAD⁺ concentrations compared to adipose tissue.
Is it necessary to add stabilizers to my samples before shipping?
Depending on the matrix, certain stabilizers (e.g., acidification) may help preserve NAD⁺/NADH ratios. Our team can advise you on suitable additives for your specific sample type.
Can you measure isotopically labeled NAD⁺/NADH for tracer studies?
Yes. Our LC-MS/MS methods can detect both natural and labeled isotopologues, making our service suitable for metabolic flux analysis and stable isotope tracing experiments.
Do temperature fluctuations during shipping impact Coenzyme I measurements?
They can. Repeated freeze-thaw cycles and elevated temperatures may degrade NAD⁺ and NADH. We recommend shipping samples on dry ice and minimizing temperature changes.
Can I submit historical samples stored for a long period?
Potentially. However, NAD⁺ and NADH can degrade over time, even at -80°C. It’s best to discuss the storage history with us so we can assess the feasibility of reliable analysis.
Can you detect NAD⁺ precursors like NMN and NR alongside Coenzyme I?
Absolutely. Our methods are validated for simultaneous detection of NAD⁺, NADH, NMN, NR, and other precursors to support studies on NAD⁺ biosynthesis and supplementation effects.
Can the method distinguish between NAD⁺ and NADH in redox ratio calculations without cross-reactivity?
Yes. Our LC-MS/MS and HPLC methods are specifically optimized to separate and quantify NAD⁺ and NADH individually, ensuring precise redox ratio assessments without analytical overlap.
Can you handle samples containing interfering substances like high salt or detergent levels?
We can usually adapt extraction and cleanup steps to remove interfering substances. However, discussing sample composition beforehand helps ensure method compatibility and accurate results.
Learn about other Q&A about proteomics technology.
Publications
Here are some of the metabolomics-related papers published by our clients:

- Methyl donor supplementation reduces phospho‐Tau, Fyn and demethylated protein phosphatase 2A levels and mitigates learning and motor deficits in a mouse model of tauopathy. 2023. https://doi.org/10.1111/nan.12931
- A human iPSC-derived hepatocyte screen identifies compounds that inhibit production of Apolipoprotein B. 2023. https://doi.org/10.1038/s42003-023-04739-9
- The activity of the aryl hydrocarbon receptor in T cells tunes the gut microenvironment to sustain autoimmunity and neuroinflammation. 2023. https://doi.org/10.1371/journal.pbio.3002000
- Lipid droplet-associated lncRNA LIPTER preserves cardiac lipid metabolism. 2023. https://doi.org/10.1038/s41556-023-01162-4
- Inflammation primes the kidney for recovery by activating AZIN1 A-to-I editing. 2023. https://doi.org/10.1101/2023.11.09.566426
- Anti-inflammatory activity of black soldier fly oil associated with modulation of TLR signaling: A metabolomic approach. 2023. https://doi.org/10.3390/ijms241310634
- Plant Growth Promotion, Phytohormone Production and Genomics of the Rhizosphere-Associated Microalga, Micractinium rhizosphaerae sp. 2023. https://doi.org/10.3390/plants12030651