- Service Details
- FAQ
- Publications
Proteins found in nature vary in size from 5 kDa to greater than 400 kDa. Protein digestion presents the process to cut proteins into shorter fragments, known as peptides. It allows for the identification and characterization of proteins according to their properties. Protein digestion is a crucial step prior to mass spectrometry (MS) analysis of peptides for successful protein identification and characterization, biomarker discovery, and systems biology. Although it is possible for MS to study intact proteins, the smaller peptides facilitate protein identification and improve the coverage of proteins that might be reduced due to solubility and heterogeneity. Therefore, the most common proteomic approaches utilize site-specific digestion to generate smaller fragments. Peptides are easier for separation and characterization by high performance liquid chromatography (HPLC) and HLPC-coupled MS.
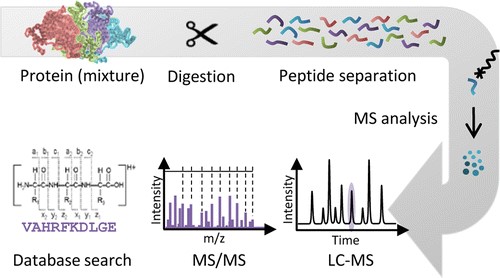
Figure 1. The general workflow of MS analysis.
Protein Digestion Services at Creative Proteomics
We offer two advanced protein digestion methods tailored to meet diverse research needs:
- In-Gel Digestion: This method involves digesting proteins directly within a polyacrylamide gel matrix. It is ideal for proteins separated by 1-D or 2-D electrophoresis. In-gel digestion is known for its robustness, effectiveness, and reproducibility. Despite these advantages, it is a labor-intensive and time-consuming process.
- In-Solution Digestion: This approach starts with protein precipitation using chloroform/methanol, followed by resolubilization in urea and subsequent digestion with trypsin or other proteases. This method is generally quicker than in-gel digestion but may result in some sample loss due to incomplete resolubilization of aggregated proteins.
Both methods are designed to efficiently solubilize hydrophobic proteins, digest proteins with trypsin or other proteases, and remove SDS, ensuring high-quality results for your proteomics needs.
Workflow of Protein Digestion
At Creative Proteomics, we provide both in-gel and in-solution protein digestion services for protein analysis. The workflow of protein digestion consists of the following steps:
1. Lysate preparation. Lysis, fractionation, depletion, enrichment, and dialysis.
2. In-solution or in-gel digestion.
a. Protein denaturation using chaotropic agents such as urea and guanidine;
b. Reduction of disulfide bridges using DTT;
c. Alkylation of the cysteines by iodoacetic acid or iodoacetamide;
d. Remove regents and exchange buffer;
e. Overnight denaturation with trypsin or other proteases in an ammonium bicarbonate buffer at suitable pH and temperature for about 18 h;
f. Stop the digestion by the addition of (formic) acid.
3. Protein enrichment/cleanup.
We also provide the high-throughput, automated protein digestion service with 96-well formats.
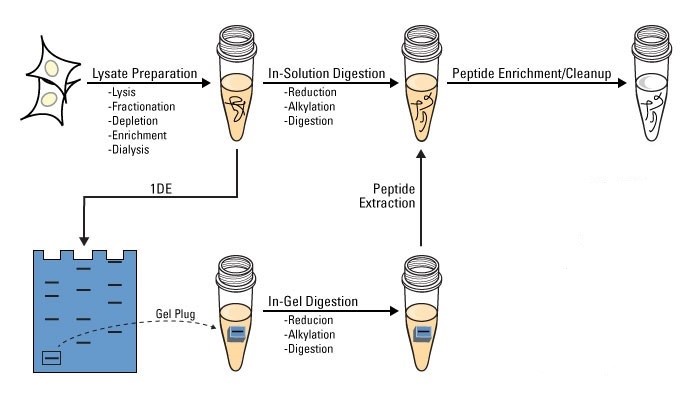
Figure 2. The workflow of protein digestion.
Sample Requirement
We accept a range of samples, such as:
- Native proteins from natural sources
- Fusion and tagged proteins from recombinant sources
- Antibodies of different isotypes
- Membrane proteins
Please submit at least 100 ug of protein. For proteins in solution, the concentration should be greater than 1 mg/mL.
Advantages of Protein Digestion
- Flexibility. We provide both in-gel and in-solution protein digestion strategies.
- Accuracy and improved coverage. Our digestion service can greatly improve the accuracy and coverage of MS analysis.
- Multiple applications. Facilitate protein identification and characterization, discover disease biomarkers, and contribute to the understanding of biological processes.
- Complete proteomics services. We provide complete proteomics services that cover the whole process of proteomics analysis.
Creative Proteomics provides in-gel and in-solution protein digestion services for successful protein and proteome analysis. As every project has different requirements, please contact our specialists to discuss your specific needs.
Reference
- Switzar L, Giera M, Niessen W M A. Protein digestion: an overview of the available techniques and recent developments. Journal of proteome research, 2013, 12(3): 1067-1077.
What are the benefits of in-gel digestion?
Resolution and Specificity: It allows for precise analysis of proteins that have been separated into distinct bands or spots on a gel. This high-resolution separation helps in identifying individual proteins and characterizing their properties.
Reduced Complexity: By digesting proteins directly within the gel, you avoid issues related to sample handling and potential losses during transfer and processing.
Reproducibility: The process is well-established and reproducible, making it a reliable choice for detailed proteomics studies.
What are the advantages of in-solution digestion?
Speed and Efficiency: It is generally faster than in-gel digestion as it bypasses the gel electrophoresis step, making it suitable for larger sample numbers or when time is a constraint.
Flexibility: This method is versatile and can be used with a wide range of sample types, including those not separated by electrophoresis.
Ease of Implementation: The method involves fewer steps compared to in-gel digestion, which can simplify the workflow.
Are there any limitations to these methods?
Each method has its limitations:
- In-Gel Digestion: While effective, this method is labor-intensive and time-consuming. It requires careful handling of gel slices and may be less practical for processing a large number of samples simultaneously.
- In-Solution Digestion: This approach may result in sample losses due to incomplete resolubilization of proteins, especially when dealing with proteins that aggregate or are poorly soluble in the urea solution. There can also be issues with contaminants if the initial precipitation step is not thorough.
How do I choose between in-gel and in-solution digestion?
Choosing the appropriate method depends on your experimental requirements:
In-Gel Digestion: Opt for this method if you have proteins that have been separated by gel electrophoresis and you need high-resolution analysis of specific bands or spots.
In-Solution Digestion: This method is preferable if you are dealing with complex mixtures that have not been previously separated or if you need a quicker, more streamlined digestion process.
Can you handle complex samples with these digestion methods?
Yes, both methods are designed to handle complex samples. In-gel digestion works well for samples that have been separated and need detailed analysis, while in-solution digestion can be applied to complex mixtures where separation is not a prerequisite. However, the effectiveness of digestion and the quality of data obtained can vary based on the sample type and the chosen method.
How do you ensure the quality and reproducibility of your digestion services?
We implement stringent quality control measures throughout the digestion process:
- Standard Operating Procedures: We follow well-established protocols to ensure consistency and accuracy.
- Experienced Personnel: Our skilled team monitors each step, from sample preparation to digestion and analysis, to maintain high standards.
- Quality Checks: We perform regular checks and validations to ensure that the digestion process yields reliable and reproducible results.
What should I provide for the digestion service?
For optimal results, please provide:
- Sample Information: Details about your protein samples, including concentration, buffer conditions, and any special handling requirements.
- Sample Format: Samples should be in a format compatible with the chosen digestion method (e.g., gel slices for in-gel digestion or solution for in-solution digestion).
- Specific Instructions: Any particular requirements or preferences for your study, such as desired protease or digestion conditions.
What types of proteins are suitable for each digestion method?
In-Gel Digestion: This method is suitable for proteins that have been separated by electrophoresis and are well-resolved into bands or spots. It is ideal for analyzing individual proteins in a complex mixture.
In-Solution Digestion: This method is more suitable for crude or complex protein mixtures that have not been separated. It works well for a wide range of proteins, including those that may aggregate or are otherwise challenging to handle.
How do you handle protein samples with high levels of contaminants or interfering substances?
For samples with high levels of contaminants, we perform additional purification steps before digestion. In the in-solution digestion method, we use chloroform/methanol precipitation to remove contaminants and concentrate the proteins. For in-gel digestion, we ensure that gel slices are thoroughly washed to remove residual contaminants. If necessary, we can employ additional clean-up procedures to ensure high-quality results.
What is the expected yield of peptides after digestion?
The yield of peptides can vary depending on the protein's size, solubility, and the digestion method used. Generally, you can expect a high yield of peptides if the digestion process is optimized and if the proteins are well-solubilized. In-solution digestion typically results in a higher peptide yield compared to in-gel digestion, due to fewer losses during sample handling.
Can you perform digestion on post-translationally modified proteins?
Yes, we can handle proteins with post-translational modifications (PTMs). Both in-gel and in-solution digestion methods are compatible with PTMs, although the choice of digestion conditions might be adjusted to optimize the recovery and analysis of modified peptides.
Untargeted proteomics and stage-specific Huntington's disease reveal biological pathways, and potential protein biomarkers.
Papanicolaou, Eleni Zamba, Christiana Christodoulou, and Christiana Demetriou.
Journey: Research Square
Year: 2024
https://doi.org/10.21203/rs.3.rs-4508811/v1
Immunoproteomics characterization of Leishmania panamensis proteins for potential clinical diagnosis of mucosal Leishmaniasis.
Caraballo‐Guzmán, A. J., Ospina‐Villa, J. D., Cuesta‐Caicedo, A. P., & Sánchez‐Jiménez, M. M.
Journey: Parasite Immunology
Year: 2021
https://doi.org/10.1111/pim.12824














