Anthocyanins Analysis Service
At Creative Proteomics, we provide cutting-edge anthocyanin analysis for academia and industry. Utilizing advanced UPLC-QTOF-MS and triple quadrupole platforms, we deliver:
- Precise Quantification
- Structural Characterization
- Metabolic Pathway Mapping
Our data-driven solutions support food scientists, agricultural researchers, and nutraceutical developers in:
- Optimizing product quality
- Validating bioactive content
- Accelerating breeding programs
Gain deeper insights into anthocyanin stability, biosynthesis, and degradation dynamics — with accuracy you can trust.
Submit Your Request Now
×- What We Provide
- Advantage
- Workflow
- Technology Platform
- Sample Requirements
- FAQ
- Case Study
What are Anthocyanins?
Anthocyanins are water-soluble flavonoid pigments responsible for the red, purple, and blue colors in many fruits, vegetables, and flowers. Beyond their role in plant physiology—where they contribute to photoprotection and stress resistance—anthocyanins are valued for their antioxidant, antimicrobial, and anti-inflammatory properties. Structurally, they are glycosylated derivatives of anthocyanidins and are commonly found in acylated and non-acylated forms. The complexity of anthocyanin profiles, combined with their sensitivity to pH, light, and temperature, requires advanced analytical techniques for accurate identification and quantification.
Why Analyze Anthocyanins?
Anthocyanin analysis plays a critical role in multiple scientific and commercial contexts:
- Functional Ingredient Profiling: Evaluate anthocyanin concentration in plant-based products to determine health-promoting properties.
- Cultivar Differentiation: Discriminate between varieties based on their anthocyanin biosynthetic pathways.
- Food Quality Assurance: Assess anthocyanin stability during processing and storage.
- Plant Stress Monitoring: Track anthocyanin production under biotic or abiotic stress conditions.
- Agricultural Breeding Programs: Identify high-anthocyanin lines through metabolic screening.
Anthocyanins Analysis Service Offered by Creative Proteomics
- Targeted Anthocyanin Quantification: Precise quantification of major anthocyanins (e.g., cyanidin-3-glucoside, malvidin-3-galactoside) using internal standards and LC-MS/MS. Ideal for comparative studies and formulation consistency.
- Untargeted Anthocyanin Profiling (Untargeted Metabolomics): Broad-spectrum profiling to identify known and novel anthocyanin compounds using high-resolution LC-QTOF-MS. Suitable for discovery research and chemotaxonomic studies.
- Structural Characterization of Anthocyanin Derivatives: In-depth identification of glycosylation and acylation patterns through tandem mass spectrometry. Supports novel compound validation and structural annotation.
- Stability and Degradation Analysis: Monitoring anthocyanin degradation under variable pH, temperature, and light conditions. Supports shelf-life estimation, packaging validation, and formulation optimization.
- Biosynthetic Pathway Mapping: Analysis of anthocyanin precursors, intermediates, and byproducts (e.g., phenylalanine, chalcone, flavanones). Enables metabolic flux tracing and gene-pathway linkage.
- Comparative Analysis Across Genotypes or Treatments: Statistical comparison of anthocyanin profiles across different cultivars, environments, or treatment groups using PCA, clustering, and multivariate analysis.
- Custom Method Development: Tailored analytical method creation for unique sample types or rare anthocyanins, including co-development services for specialized research projects.
Detected Anthocyanins and Related Analytes
| Category | Representative Compounds |
|---|---|
| Major Anthocyanins | Cyanidin, Delphinidin, Pelargonidin, Peonidin, Malvidin, Petunidin |
| Glycosylated Anthocyanins | Cyanidin-3-glucoside, Delphinidin-3-sambubioside, Malvidin-3-galactoside, Peonidin-3-rutinoside |
| Acylated Anthocyanins | Cyanidin-3-(6''-malonylglucoside), Delphinidin-3-(p-coumaroylglucoside), Petunidin-3-caffeoylglucoside |
| Anthocyanin Degradation Products | Protocatechuic acid, Gallic acid, Syringic acid, Vanillic acid |
| Flavonoid Intermediates | Naringenin, Dihydrokaempferol, Dihydroquercetin, Leucocyanidin, Eriodictyol |
| Biosynthetic Precursors | Phenylalanine, Cinnamic acid, p-Coumaric acid, Ferulic acid, Sinapic acid |
| Pathway-Related Enzymatic Products | Chalcone, Chalcone isomerase (CHI) intermediates, Flavanone, Flavonol |
| Additional Phenolic Compounds | Quercetin, Kaempferol, Myricetin, Luteolin, Apigenin |
| Other Detected Polyphenols | Resveratrol, Catechin, Epicatechin |
Advantages of Anthocyanins Assay
- Detection limit as low as 1 ng/mL for major anthocyanins using UPLC-MS/MS.
- Quantification accuracy > 95% with internal standard normalization.
- Retention time precision with RSD < 2% across technical replicates.
- Method reproducibility with inter-batch variation < 5% (validated over 3 runs).
- Comprehensive coverage of over 200 anthocyanins and related metabolites.
- High-throughput capability up to 200 samples/day using automated workflow.
- Mass accuracy within ±2 ppm for structural identification via QTOF-MS.
- Peak resolution > 98% for closely eluting anthocyanin isomers.
- Pathway mapping supported by over 1,000 curated biosynthetic entries.
- Data turnaround time as fast as 10 business days from sample receipt.
Workflow for Anthocyanins Analysis Service
Technology Platform for Anthocyanins Analysis Service
Agilent 6495C Triple Quadrupole LC/MS: Targeted quantification of anthocyanins and derivatives at ultra-trace levels.
- Detection limit as low as 1 ng/mL.
- High selectivity for structurally similar anthocyanins.
- Multiple reaction monitoring (MRM) mode for high-precision quantification.
- Ideal for comparing anthocyanin levels across cultivars, conditions, or formulations.
Agilent 1260 Infinity II HPLC with Diode Array Detection (DAD): Chromatographic separation and spectral profiling of anthocyanins.
- Retention time reproducibility with <2% RSD.
- UV-Vis detection at 520–540 nm for anthocyanin-specific absorption.
- Effective for distinguishing isomers and monitoring stability during processing or storage.

Agilent 6495C Triple Quadrupole (Figure from Agilent)

Agilent 1260 Infinity II HPLC (Fig from Agilent)
Sample Requirements for Anthocyanins Analysis Service
| Sample Type | Required Amount | Notes |
|---|---|---|
| Plant Tissue (leaf, root, stem, etc.) | ≥ 200 mg (fresh weight) / ≥ 50 mg (dry weight) | Flash-freeze in liquid nitrogen and store at -80°C; avoid repeated freeze-thaw. |
| Fruits & Vegetables | ≥ 200 mg (fresh) | Homogenize thoroughly; avoid browning or oxidation prior to submission. |
| Fermented Products (wine, tea, etc.) | ≥ 1 mL | Store at -20°C in sealed vials; avoid light exposure. |
| Food Samples (powder, extract) | ≥ 100 mg | Provide dry powder or concentrated extract; indicate solvent if applicable. |
| Serum / Plasma | ≥ 200 µL | Collect using EDTA or heparin tubes; centrifuge and aliquot; freeze at -80°C. |
| Urine | ≥ 1 mL | Collect midstream, store at -80°C; avoid preservatives. |
| Cell / Microbial Pellet | ≥ 1 × 10⁷ cells | Wash with PBS and freeze at -80°C promptly after collection. |
| Culture Supernatant | ≥ 1 mL | Filter sterilize if needed; avoid contamination. |
FAQ of Anthocyanins Analysis Service
What is the minimum sample quantity required for analysis?
Typically, 10–50 mg of plant tissue or 1–5 mL of liquid samples (e.g., juice, wine). We optimize protocols for low-yield samples.
How do you handle anthocyanin degradation during shipping?
Ship samples on dry ice or in stabilizing buffers (provided upon request). Avoid light exposure using amber vials.
Can your analysis distinguish between anthocyanin degradation products and native compounds?
Yes, our LC-MS/MS methods separate and quantify degradation markers (e.g., protocatechuic acid) from intact anthocyanins.
Do you support data interpretation for agricultural studies?
We provide statistical correlations between anthocyanin profiles and environmental/growth conditions, including heatmaps and pathway activity scores.
How do I choose between targeted quantification and untargeted profiling?
Targeted is ideal for validating known anthocyanins (e.g., quality control). Untargeted suits novel compound discovery or pathway studies.
Can I combine anthocyanin analysis with other polyphenol quantification?
Yes, our metabolomics platform supports simultaneous profiling of anthocyanins, flavonols, catechins, and phenolic acids.
What software tools do you provide for data visualization?
Interactive PCA plots, volcano plots, and pathway maps via MetaboAnalyst 5.0. Raw data formats (mzML, CSV) are also available.
Do you analyze anthocyanins in processed foods with additives?
Yes, but preservatives like sulfites may interfere. Pre-treatment protocols can minimize matrix effects—contact us for details.
How are results delivered?
Secure online portal access with downloadable PDF reports, Excel files, and annotated chromatograms/spectra.
Can your service identify anthocyanin-binding proteins or complexes?
No—this requires separate proteomics workflows. We focus on free and conjugated anthocyanins in extracts.
Learn about other Q&A.
Anthocyanins Analysis Service Case Study

Title: Generation of Purple-Violet Chrysanthemums via Anthocyanin B-Ring Hydroxylation and Glucosylation Introduced from Osteospermum hybrid F3'5'H and Clitoria ternatea A3'5'GT
Journal: Ornamental Plant Research
Published: 2021
- Background
- Methods
- Results
- Conclusion
- Reference
Chrysanthemums naturally lack the ability to synthesize delphinidin, the blue anthocyanin pigment, due to the absence of the F3'5'H gene—an essential hydroxylase in the flavonoid biosynthetic pathway. As a result, blue or blue-purple chrysanthemum flowers do not occur naturally. Previous approaches to overcome this limitation have involved introducing exogenous F3'5'H genes from various ornamental plants, combined with glycosylation-related genes, to enable delphinidin accumulation and produce novel flower colors. This study investigates the co-expression of Osteospermum hybrid F3'5'H (OhF3'5'H) and Clitoria ternatea A3'5'GT, alongside a site-directed mutant of the native CmF3'H (T485S), to promote delphinidin biosynthesis and generate purple-violet chrysanthemum flowers.
Chrysanthemum cultivar 'Nannong Fencui' was genetically transformed using Agrobacterium tumefaciens EHA105 carrying recombinant binary vectors. Leaf explants underwent co-cultivation, selection, and regeneration to obtain kanamycin-resistant transgenic lines expressing flavonoid biosynthesis genes.
Total RNA was extracted from ray florets of transgenic and wild-type plants for cDNA synthesis and RT-PCR validation. Flower color phenotypes were evaluated using the RHS Colour Chart and Lab* color space measurements.
For pigment profiling, ray florets were ground in liquid nitrogen, extracted using an acidified methanol solvent system, and analyzed by UPLC-MS/MS. Separation was performed on a BEH C18 column, and compounds were detected using positive ESI mode. Mass spectrometry data acquisition and processing were performed using MassLynx and UNIFI software, enabling qualitative and quantitative analysis of anthocyanin composition.
Creative Proteomics Services
Based on this study, Creative Proteomics can provide Anthocyanins Analysis Services supporting:
- Targeted extraction of anthocyanins and related flavonoids from plant tissues.
- UPLC-QToF-MS/MS analysis for high-resolution detection and identification.
- Quantitative profiling of anthocyanin compounds to reveal pigment composition changes in transgenic versus wild-type lines.
Transgenic chrysanthemum lines expressing OhF3'5'H or mutant CmF3'Hm along with CtA3'5'GT were successfully obtained. Expression of target genes was confirmed by RT-PCR. Flower color in transgenic lines shifted visibly compared to wild-type: OhF3'5'H-expressing lines (OH9-3, OH23-8) exhibited light purple to violet tones, while CmF3'Hm lines (Cm-29, Cm-38) showed paler purples.
Colorimetric analysis revealed both CmF3'Hm and OhF3'5'H lines had reduced red and blue chromaticity versus wild-type. However, only OhF3'5'H lines exhibited higher blue chromaticity and a shift toward blue hue.
UPLC-ESI-MS/MS analysis showed that:
- Wild-type lines accumulated cyanidin-based anthocyanins A1 and A2.
- CmF3'Hm lines also contained only cyanidin derivatives (A3, A4), indicating no enzymatic shift to delphinidin pathway.
- OhF3'5'H lines accumulated both cyanidin (A3, A4) and delphinidin derivatives (A7, A8, A9), confirming OhF3'5'H function in delphinidin biosynthesis.
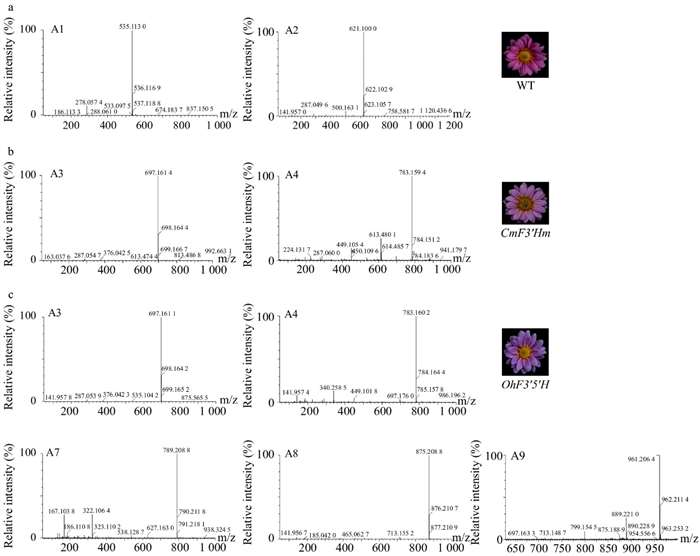 Mass spectrometric analysis results of anthocyanins of transgenic and wild-type chrysanthemums.
Mass spectrometric analysis results of anthocyanins of transgenic and wild-type chrysanthemums.
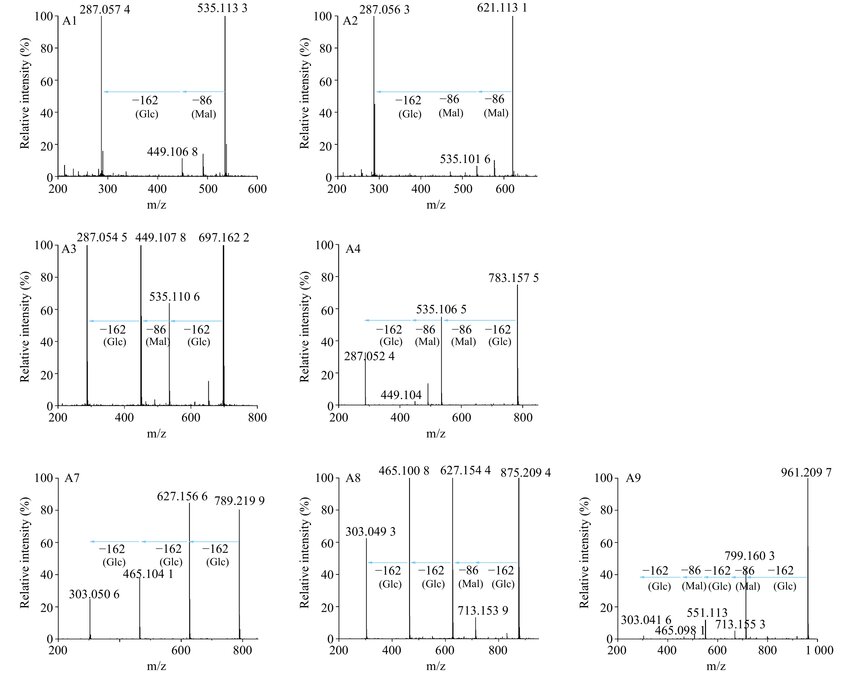 MS/MS analysis of anthocyanins in transgenic and wild-type chrysanthemums.
MS/MS analysis of anthocyanins in transgenic and wild-type chrysanthemums.
Expression of OhF3'5'H enables delphinidin biosynthesis and results in a blue-shifted flower color in chrysanthemum, while mutant CmF3'Hm fails to induce this effect. Anthocyanin profiling confirms the production of delphinidin-based pigments only in OhF3'5'H lines, validating its functional role in blue coloration.
Reference
- Han, Xiaoying, et al. "Generation of purple-violet chrysanthemums via anthocyanin B-ring hydroxylation and glucosylation introduced from Osteospermum hybrid F3'5'H and Clitoria ternatea A3'5'GT." Ornamental Plant Research 1.1 (2021): 1-9. https://doi.org/10.48130/OPR-2021-0004
Publications
Here are some publications in Metabolomics research from our clients:

- A human iPSC-derived hepatocyte screen identifies compounds that inhibit production of Apolipoprotein B. 2023. https://doi.org/10.1038/s42003-023-04739-9
- WI12 Rhg1 interacts with DELLAs and mediates soybean cyst nematode resistance through hormone pathways. 2022. https://doi.org/10.1111/pbi.13709
- Pan-lysyl oxidase inhibition disrupts fibroinflammatory tumor stroma, rendering cholangiocarcinoma susceptible to chemotherapy. 2024. https://doi.org/10.1097/HC9.0000000000000502
- Sex modifies the impact of type 2 diabetes mellitus on the murine whole brain metabolome. 2023. https://doi.org/10.3390/metabo13091012
- Lipin-1 regulates lipid catabolism in pro-resolving macrophages. 2020. https://doi.org/10.1101/2020.06.03.121293







