Palmitoylation Analysis Service
Creative Proteomics offers specialized services for palmitoylated protein detection, supported by:
- Advanced platforms: LC-MS/MS, click chemistry, and enrichment methods
- Expert guidance: From experimental design to data analysis
- Flexible service packages: Suitable for academic and industrial needs
Our full protein PTM analysis portfolio also includes phosphorylation, acetylation, and glycosylation studies.
Submit Your Request Now
×- Introduction
- How to Solve the Challenges
- Service Overview
- Advantage
- Sample Requirement
- Case Study
- FAQ
- Publication
What is Protein Palmitoylation?
When it comes to fine-tuning protein function, post-translational modifications are key players. Among the more common types—like phosphorylation and glycosylation—S-palmitoylation stands out for its reversibility and its powerful role in controlling how proteins behave inside cells.
This unique process involves the attachment of fatty acids, typically palmitic acid, to specific amino acids on proteins. By increasing a protein's hydrophobicity, palmitoylation influences everything from its cellular destination to its role in signalling pathways. For scientists looking to optimise monoclonal antibody production, understanding this mechanism can open new doors in therapeutic development.
Why Analyze Protein Palmitoylation?
Unlike permanent modifications, palmitoylation is reversible, making it a dynamic regulator of protein function. This flexibility allows cells to rapidly respond to environmental changes by altering protein activity, location, or interactions.
Key impacts of palmitoylation include:
- Protein trafficking: It helps shuttle proteins to specific organelles.
- Membrane association: Increases lipid affinity, anchoring proteins where they're needed.
- Signal transduction: Fine-tunes key pathways involved in growth, immunity, and cell death.
Understanding where palmitoylation happens in a protein helps researchers:
- Track how proteins behave in different environments
- Discover how signaling pathways work
- Identify new drug targets
This makes palmitoylation site identification essential for fields like oncology, virology, and neurobiology. By knowing how and where this modification occurs, scientists can uncover hidden mechanisms in diseases—and even potential ways to treat them.
Beyond S-palmitoylation: A Broader Landscape
While S-palmitoylation (a form of S-acylation) is the best-studied, other types also exist:
N-palmitoylation: Palmitic acid links to amino groups on glycine, cysteine, or lysine.
O-palmitoylation: Occurs via hydroxy groups on serine or threonine residues.
Each variant has distinct biochemical consequences and could represent new levers in drug development or disease modelling.
The Roadblocks in Palmitoylation Research
Despite growing interest, studying palmitoylation has long been a slow and resource-heavy process. Traditional methods—like site-directed mutagenesis or chemical inhibitors—have helped confirm its presence. But compared to better-understood modifications like phosphorylation or glycosylation, progress has lagged behind.
This gap limits insights into how palmitoylation shapes protein function, especially in drug design or monoclonal antibody production. Fortunately, newer technologies are changing the game.
How We Solve Common Detection Challenges
Creative Proteomics addresses these with:
- Optimised sample preparation to preserve labile modifications
- Selective chemistry for targeted enrichment
- Robust data analysis pipelines that filter false positives and validate results across replicates
Mass Spectrometry and Proteomics Lead the Way
Advancements in mass spectrometry (MS) and proteomic tools have pushed palmitoylation research forward. Scientists can now directly detect modified proteins and map palmitoylation sites with greater speed and accuracy.
Key breakthroughs include:
ABE (Acyl-Biotin Exchange): Enables site-specific identification of palmitoylated residues.
Metabolic labeling: Uses palmitate analogs to tag and trace modified proteins inside live cells.
These techniques have uncovered a growing list of palmitoylated targets, helping researchers connect specific modifications to biological functions.
High-Throughput Discovery with Click Chemistry
More recently, click chemistry has emerged as a powerful companion to ABE. When combined with high-throughput protein profiling, it allows researchers to scan for palmitoylation events across a wide range of tissues and even clinical samples.
Together, these methods provide a scalable way to:
Identify novel palmitoylation targets
Quantify dynamic changes in response to drugs
Support biomarker discovery in disease research
For pharma teams seeking monoclonal antibody production tips, mastering these tools could streamline development by revealing new control points in protein function.
Comparison of Protein Palmitoylation Detection Methods
| Method | Principle | Advantages | Limitations |
|---|---|---|---|
| Click Chemistry | Azide-alkyne reaction for labeling | High sensitivity, non-radioactive | May interfere with cellular metabolism |
| Biotin Exchange | Biotinylation after thioester cleavage | Quantitative, cysteine-specific | Complex workflows, potential artifacts |
| LC-MS/MS | Detects mass shift of modified proteins | High accuracy, high throughput | Requires specialized equipment |
Creative Proteomics' Palmitoylation Site Identification Service
Accurately identifying palmitoylation sites is essential for understanding how proteins behave in cells—especially when developing precision biologics or improving monoclonal antibody production workflows. Our lab offers two complementary service platforms that combine cutting-edge biochemistry with high-resolution proteomics.
Workflow 1: Click Chemistry-Based Detection
1. Metabolic Labeling
- Target proteins are labeled with palmitic acid analogs during live-cell expression.
2. Click Chemistry Reaction
- A catalyst-driven click reaction attaches biotin tags to the palmitoylated residues.
3. Affinity Enrichment
- Streptavidin beads isolate the biotin-labeled peptides.
4. Mass Spectrometry (LC-MS/MS)
- Advanced LC-MS/MS is used to identify and characterise the modified peptides.
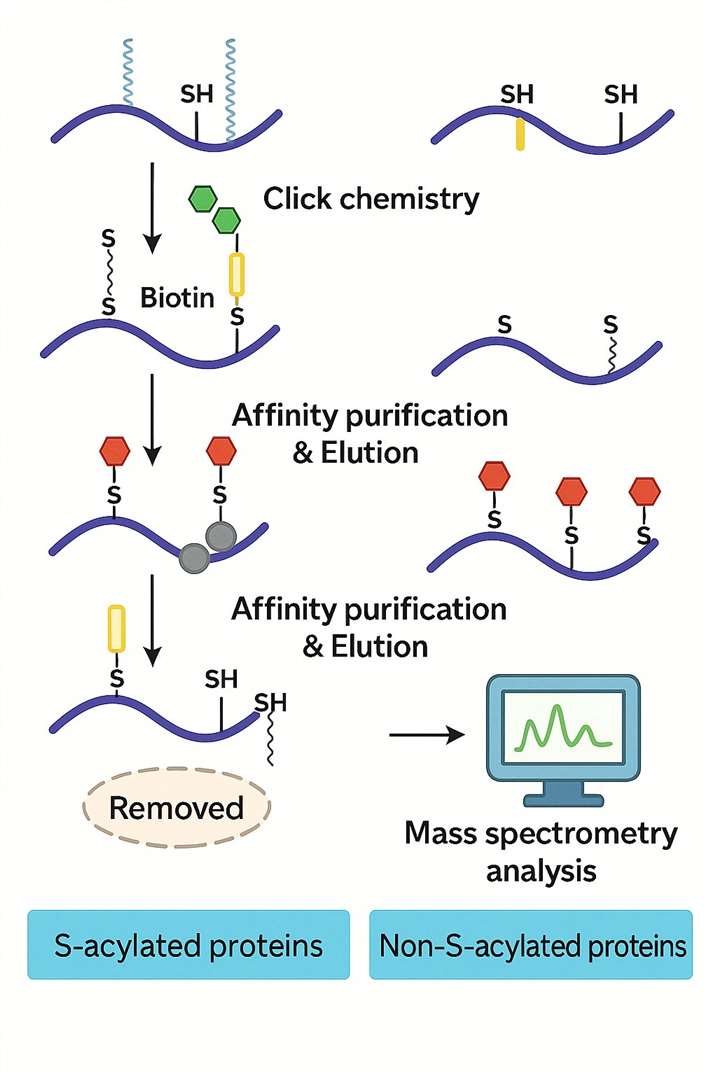 This approach allows dynamic profiling of palmitoylation under different conditions or treatments.
This approach allows dynamic profiling of palmitoylation under different conditions or treatments.
Workflow 2: Biotin Exchange (ABE) Method
Ideal for high-sensitivity detection, ABE provides a robust chemical strategy for site-specific analysis:
1. Free Thiol Alkylation
- Native cysteine thiol groups are first blocked to prevent background noise.
2. Selective Depalmitoylation
- Palmitoyl groups are cleaved, exposing formerly modified sites.
3. Biotin Tagging
- Newly revealed thiols are labelled with biotin.
4. Affinity Enrichment
- Biotinylated peptides are captured using streptavidin beads.
5. Mass Spectrometry (LC-MS/MS)
- LC-MS/MS identifies and maps palmitoylation sites with high confidence.
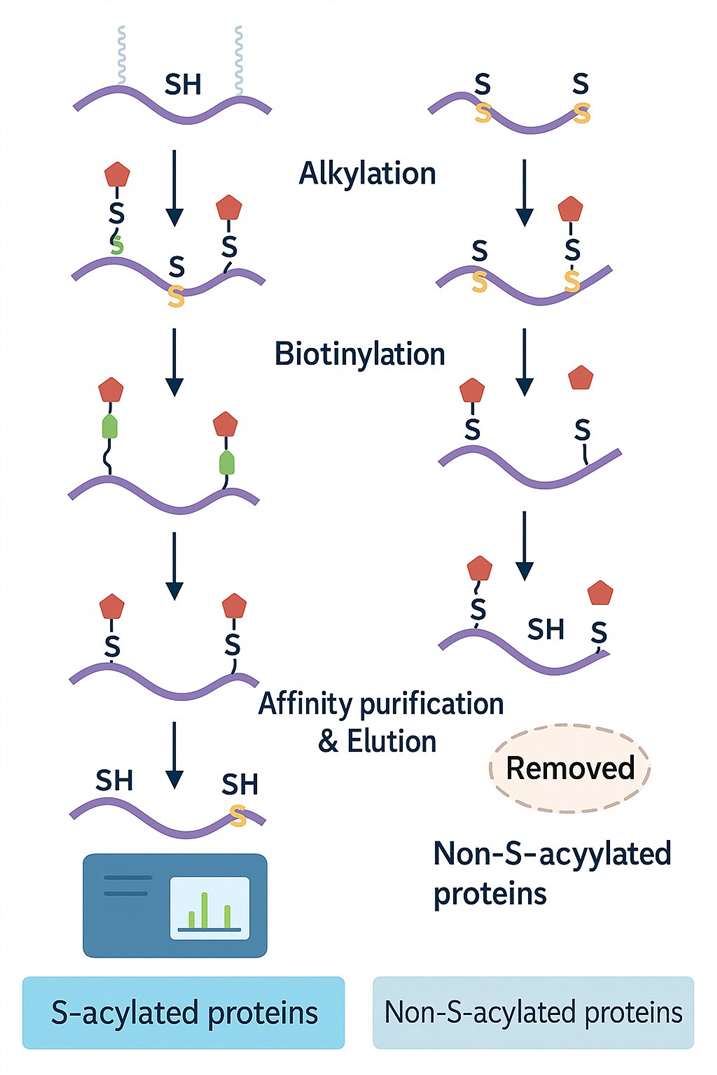
Why Use Both?
While click chemistry excels at live-cell tracking, ABE offers higher sensitivity for endpoint analysis. Using both can yield a comprehensive palmitoylation profile across biological states or tissue types.
Service Advantages
- Easy and simple operation: no isotope labeling required; simple experimental procedure
- High precision and accuracy: specific modification sites can be identified and analyzed; the Thermo Fisher Mass Spectrometer can be used to obtain additional peptide information and modification sites
- Wide range of detection: palmitoylation sites can be detected in various cells or tissues
Sample Requirements
| Sample Type | Sample Amount | Additional Notes |
|---|---|---|
| Fresh animal tissue | ≥600 mg | |
| Fresh plant tissue | ≥6 g | |
| Cell culture | ≥1×10⁷ cells/tube × 3 tubes | |
| Fungi and bacteria | ≥600 mg | |
| Serum, plasma | 450 μL × 4 tubes | |
| Protein solution | Total protein of 5–10 mg | |
| Body fluid – Urine | 15 mL × 4 tubes | Centrifuge at 1000 × g for 5 minutes and discard sediment |
| Body fluid – Others (saliva, amniotic fluid, cell culture supernatant, etc.) | >15 mL |
Case Study

Protein S-palmitoylation modification: implications in tumor and tumor immune microenvironment
- Overview
- Methods Used in This Study
- Key Findings
Protein S-palmitoylation is a reversible lipid modification where a palmitoyl group is covalently attached to cysteine residues. This biochemical switch controls a protein's localisation, stability, and function. In oncology, S-palmitoylation regulates critical pathways such as oncogene activation, tumour suppressor inactivation, immune evasion, metabolic reprogramming, and programmed cell death.
Recent mass spectrometry-based proteomic studies have identified numerous palmitoylated proteins in both tumour and immune cells. These findings shed light on how S-palmitoylation shapes the tumour immune microenvironment—a growing area of interest in immuno-oncology research.
To decode the palmitoylation landscape, the following advanced techniques were employed:
Mass Spectrometry (MS):
Using high-resolution MS combined with acyl-biotin exchange (ABE) and palmitate analogue labelling, researchers systematically profiled palmitoylated proteins across tumour and immune cell models.
Quantitative proteomics was used to assess how S-palmitoylation affects protein activity, signal transduction, and cell behaviour.
Software-based enrichment and pathway mapping tools helped functionally annotate palmitoylated proteins and reveal their roles in cancer progression and immune regulation.
S-palmitoylation plays diverse and critical roles within the tumour microenvironment:
Modulates Cancer Signalling
By stabilising and localising key proteins like EGFR, CDCP1, AKT, and mTOR, S-palmitoylation enhances pro-tumour signalling. This modification promotes cell proliferation and metastasis in various cancer models.
Stabilises Immune Checkpoint Proteins
S-palmitoylation increases the half-life of PD-L1 and PD-1 by protecting them from degradation. This contributes to immune escape. Inhibiting palmitoylation of these targets has shown promise in boosting anti-tumour immune responses.
Regulates T Cell Function
T cell receptor (TCR) signalling relies on palmitoylated proteins like LCK, LAT, and PLC-γ1. These modifications modulate T cell activation and downstream responses—highlighting a novel control point for immunotherapy.
Drives Metabolic Reprogramming
Tumour cells exploit S-palmitoylation to alter metabolic enzyme activity. Proteins like MDH2 and AK2, once palmitoylated, help rewire cellular metabolism to support tumour survival and rapid growth.
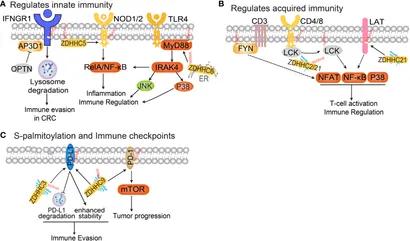 Figure 1 The functional roles of protein S-palmitoylation in anti-tumor immunity.
Figure 1 The functional roles of protein S-palmitoylation in anti-tumor immunity.
FAQs: Palmitoylation Analysis Services
Clear answers to help you get the most from your palmitoylation profiling projects.
What is protein palmitoylation?
Palmitoylation is a type of lipid modification where palmitic acid attaches to cysteine residues on a protein. This changes how the protein behaves—affecting where it goes in the cell and how it interacts with other molecules.
Why is palmitoylation important in drug development?
This reversible modification plays a major role in:
- Cell signalling and immune responses
- Protein stability and degradation
- Disease pathways like cancer, viral infections, and neurodegeneration
Understanding these changes can guide drug design and improve monoclonal antibody production strategies.
How do you detect palmitoylated proteins?
We use three core approaches:
- Click chemistry for live-cell tagging of modified proteins
- Biotin exchange assays (ABE) for precise site identification
- High-resolution LC-MS/MS for detailed mapping of modified peptides
These methods are often used together for full-spectrum analysis.
What are the main challenges in palmitoylation analysis?
Some common hurdles include:
- Its reversible nature, which can complicate detection
- Overlapping lipid modifications, like myristoylation, which require method selectivity
- Low abundance signals, demanding sensitive enrichment strategies
Our workflows are designed to address each of these issues.
What can palmitoylation data be used for?
Clients typically use this data to:
- Understand disease mechanisms at the protein level
- Identify new biomarkers for diagnostics or monitoring
- Screen therapeutic compounds that alter palmitoylation states
This kind of profiling can uncover hidden regulatory checkpoints in biologics development.
Learn about other Q&A about other technologies.
Publication
Here are some publications in PTMs proteomics research from our clients:

- Exploratory phosphoproteomics profiling of Aedes aegypti Malpighian tubules during blood meal processing reveals dramatic transition in function. Plos one. https://doi.org/10.1371/journal.pone.0271248
- The Interplay between GSK3βand Tau Ser262 Phosphorylation during the Progression of Tau Pathology. International Journal of Molecular Sciences. https://doi.org/10.3390/ijms231911610
- In Vitro Identification of Phosphorylation Sites on TcPolβ by Protein Kinases TcCK1, TcCK2, TcAUK1, and TcPKC1 and Effect of Phorbol Ester on Activation by TcPKC of TcPolβ in Trypanosoma cruzi. Microorganisms. https://doi.org/10.3390/microorganisms12050907
- HDAC4 influences the DNA damage response and counteracts senescence by assembling with HDAC1/HDAC2 to control H2BK120 acetylation and homology-directed repair. Nucleic Acids Research. https://doi.org/10.1093/nar/gkae501
References
- Amy F Roth, Junmei Wan, Aaron O Bailey, Beimeng Sun, Jason A Kuchar, William N Green, Brett S Phinney, John R Yates 3rd, Nicholas G Davis Global analysis of protein palmitoylation in yeast. Cell 2006;125(5):1003-13. https://doi.org/10.3389/fnmol.2018.00175
- Roth AF, Wan J, Bailey AO, Sun B, Kuchar JA, Green WN, Phinney BS, Yates JR 3rd, Davis NG. Global analysis of protein palmitoylation in yeast. Cell. 2006 Jun 2;125(5):1003-13. doi: 10.1016/j.cell.2006.03.042. PMID: 16751107; PMCID: PMC2246083. DOI: 10.1016/j.cell.2006.03.042












