Cardiolipin Analysis Service
Creative Proteomics offers precise cardiolipin analysis using advanced LC-MS technology, providing:
- Quantification and profiling of cardiolipin (CL), monolysocardiolipin (MLCL), and oxidized cardiolipin (OxCL).
- Insights into mitochondrial function, oxidative stress, and lipid metabolism.
- Solutions for mitochondrial dysfunction, metabolic diseases, and aging research.
- Reliable, high-throughput analysis for diverse biological samples.
Our service ensures accurate, reproducible results to support your research needs.
Submit Your Request Now
×- What We Provide
- Advantage
- Workflow
- Technology Platform
- Sample Requirements
- FAQ
- Publication
What is Cardiolipin?
Cardiolipin is a unique dimeric phospholipid predominantly located in the inner mitochondrial membrane, playing a crucial role in mitochondrial structure, bioenergetics, and apoptosis regulation. Owing to its distinctive tetra-acyl structure and tissue-specific composition, cardiolipin serves as a sensitive marker for mitochondrial function, membrane integrity, and oxidative stress response. Comprehensive analysis of cardiolipin molecular species is essential for elucidating mitochondrial dynamics, lipid metabolism, and bioenergetic performance in various biological systems.
Why Analyze Cardiolipin?
Accurate quantification and structural elucidation of cardiolipins are indispensable for high-resolution mitochondrial studies. Given its susceptibility to oxidative damage and involvement in numerous cellular pathways, alterations in cardiolipin composition can provide valuable insights into:
- Mitochondrial membrane remodeling
- Reactive oxygen species (ROS)-induced lipid peroxidation
- Mitochondrial aging and bioenergetic decline
- Lipidomic reprogramming in metabolic or degenerative conditions
- Mitochondrial biogenesis and fusion/fission dynamics
Creative Proteomics provides a comprehensive cardiolipin analysis service equipped with advanced instrumentation and standardized protocols to deliver reliable, reproducible, and highly specific lipidomic data.
Cardiolipin Analysis Service Offered by Creative Proteomics
Quantitative and Qualitative Cardiolipin Profiling
- Absolute and relative quantification of cardiolipin molecular species
- Detailed acyl chain composition analysis (e.g., CL (72:8), CL (70:5))
- Detection of minor and tissue-specific cardiolipin isoforms
Monolysocardiolipin (MLCL) and Oxidized Cardiolipin Detection
- Quantification of monolysocardiolipin (MLCL) for assessing cardiolipin remodeling
- Identification of oxidized cardiolipin (OxCL) forms under oxidative stress conditions
- Mapping of MLCL/CL ratios relevant to mitochondrial integrity
Cardiolipin Remodeling and Turnover Analysis
- Profiling of cardiolipin biosynthesis and degradation intermediates
- Assessment of remodeling enzymes' activity via substrate/product ratios
- Time-course studies for mitochondrial lipid remodeling dynamics
Metabolite and Lipid Precursor Correlation
- Measurement of cardiolipin-associated precursors: PG, CDP-DAG, PA
- Analysis of downstream metabolites related to glycerophospholipid pathways
- Isotopic labeling (optional) to trace lipid fluxes
Cardiolipin Pathway Mapping and Data Interpretation
- Integration of cardiolipin data into KEGG, HMDB, and Reactome lipid pathways
- Identification of altered cardiolipin-associated networks
- Statistical comparison across experimental groups (optional service)
Custom Assay Development (Upon Request)
- Development of targeted MRM methods for specific cardiolipin species
- Inclusion of uncommon cardiolipins from non-mammalian systems (e.g., yeast, bacteria)
- Tailored extraction protocols for low-yield or rare sample types
List of Detected Cardiolipin and Related Metabolites
| Analyte Name | Category | Description | Associated Pathway(s) |
|---|---|---|---|
| CL(72:8) | Cardiolipin | Tetra-linoleoyl cardiolipin, predominant in heart tissue | Oxidative phosphorylation, mitochondrial membrane structure |
| CL(70:5) | Cardiolipin | Mixed saturated/unsaturated chains | Electron transport chain, ROS response |
| CL(68:4) | Cardiolipin | Common mitochondrial lipid species | Mitochondrial bioenergetics |
| CL(66:2) | Cardiolipin | Saturated-rich CL species | Lipid remodeling, aging-associated changes |
| CL(72:9) | Cardiolipin | Polyunsaturated cardiolipin variant | Apoptosis and cytochrome c release |
| MLCL(54:4) | Monolysocardiolipin | Remodeling intermediate of CL | Barth syndrome marker, mitochondrial integrity |
| MLCL(52:2) | Monolysocardiolipin | Defective CL remodeling product | Lipid trafficking, mitochondrial disease |
| OxCL(72:8-OOH) | Oxidized Cardiolipin | Oxidized tetra-linoleoyl cardiolipin | ROS signaling, apoptosis regulation |
| OxCL(70:6-oxo) | Oxidized Cardiolipin | Electrophilic oxidation product | Inflammation-related lipid signaling |
| PG(34:1) | Phosphatidylglycerol | Direct precursor of CL synthesis | Glycerophospholipid metabolism |
| PG(36:2) | Phosphatidylglycerol | Mitochondrial lipid precursor | CL biosynthetic pathway |
| CDP-DAG | Cytidine diphosphate diacylglycerol | Central intermediate in CL biosynthesis | Phospholipid biosynthesis |
| PA(32:0) | Phosphatidic acid | Precursor of CDP-DAG | Glycerolipid metabolism |
| LPA(18:1) | Lysophosphatidic acid | Lipid intermediate and signaling molecule | Lipid remodeling, membrane dynamics |
| DAG(36:2) | Diacylglycerol | Intermediate in lipid turnover | PKC signaling, lipid metabolism |
| PI(38:4) | Phosphatidylinositol | Signaling lipid | Inositol phosphate metabolism |
| PS(36:1) | Phosphatidylserine | Related mitochondrial lipid | Membrane fusion/fission processes |
| PE(38:6) | Phosphatidylethanolamine | CL-interacting lipid | Membrane curvature, mitochondrial dynamics |
| LPC(18:0) | Lysophosphatidylcholine | Oxidative stress indicator | Inflammatory lipid network |
| SM(d18:1/16:0) | Sphingomyelin | Cross-pathway lipid | Sphingolipid metabolism, apoptosis |
| Cer(d18:1/24:1) | Ceramide | Mitochondrial-associated apoptosis lipid | Ceramide signaling, mitochondrial outer membrane |
Advantages of Cardiolipin Assay
- Detects cardiolipin species with an ultra-low detection limit of 1 pmol, ensuring high sensitivity for trace-level analysis.
- Enables linear quantification across five orders of magnitude with a correlation coefficient (R²) greater than 0.995.
- Covers over 200 cardiolipin and monolysocardiolipin (MLCL) species in a single analytical run.
- Delivers outstanding reproducibility, with intra-batch CV values below 5% and inter-batch CV values below 10%.
- Provides high mass accuracy with mass errors consistently controlled within ±3 ppm, ensuring reliable identification.
- Utilizes high-resolution MS platforms (≥240,000 FWHM) to separate and resolve isobaric and structurally similar species.
- Employs stable isotope-labeled internal standards for robust and absolute quantification across sample types.
- Requires minimal sample input—only 200 µL of plasma or 1×10⁷ cells for full cardiolipin profiling.
- Offers optional integrated bioinformatics reporting, including pathway enrichment analysis and statistical comparisons.
Workflow for Cardiolipin Analysis Service
Technology Platform for Cardiolipin Analysis Service
Agilent 6495C Triple Quadrupole LC-MS: Ultra-sensitive targeted quantification with femtomole-level detection and excellent reproducibility.
SCIEX Triple Quad™ 6500+: High-speed, high-throughput lipidomics platform ideal for detecting oxidized and low-abundance cardiolipin species.
Agilent 1260 Infinity II HPLC: Delivers high-efficiency chromatographic separation, enabling resolution of isomeric cardiolipins prior to MS detection.

Agilent 6495C Triple quadrupole (Figure from Agilent)

SCIEX Triple Quad™ 6500+ (Figure from Sciex)

Agilent 1260 Infinity II HPLC (Fig from Agilent)
Sample Requirements for Cardiolipin Analysis Service
| Sample Type | Minimum Quantity | Storage Conditions | Notes |
|---|---|---|---|
| Cell Pellet | ≥1×10⁷ cells | -80°C | Washed with PBS, no serum |
| Tissue | ≥50 mg | -80°C | Snap frozen, avoid repeated freeze-thaw |
| Plasma/Serum | ≥200 µL | -80°C | EDTA or heparin anticoagulants acceptable |
| Isolated Mitochondria | ≥100 µg protein | -80°C | Quantified by BCA or Bradford assay |
| Bacterial Culture | ≥10⁹ cells | -80°C | Centrifuged and washed |
| Yeast Culture | ≥5×10⁸ cells | -80°C | Standard growth media recommended |
Applications of Cardiolipin Assay Service
Mitochondrial Function Research
Cardiolipin is essential for mitochondrial energy production and stability. Its analysis aids in understanding mitochondrial dysfunction in metabolic disorders.
Lipid Metabolism
Analyze cardiolipin to investigate lipid metabolic changes in diseases.
Aging
Study age-related alterations in mitochondrial function through cardiolipin profiling.
Neurodegenerative Diseases
Investigate cardiolipin changes in Alzheimer's, Parkinson's, and other neurodegenerative conditions.
Cancer Research
Explore the role of cardiolipin in mitochondrial bioenergetics and tumor progression.
Drug Discovery
Evaluate the effects of compounds on mitochondrial health by analyzing cardiolipin.
Cardiolipin Analysis Service Case Study

Title: High-resolution mass spectrometric analysis of cardiolipin profiles in Barth syndrome
Journal: Mitochondrion
Published: 2021
- Background
- Materials & Methods
- Results
- Reference
Barth syndrome (BTHS) is a rare X-linked recessive disorder caused by mutations in the TAZ gene, leading to defects in cardiolipin (CL) remodeling in mitochondria. This results in decreased mature cardiolipin and increased monolysocardiolipin (MLCL), with variable accumulation of dilysocardiolipin (DLCL). These lipid alterations impair mitochondrial structure and function. Traditional diagnostic methods, including urinary acid analysis and TAZ gene sequencing, have limitations in sensitivity and specificity. High-resolution untargeted lipidomics offers a powerful approach to accurately identify and quantify diverse lipid species. This study aims to characterize changes in individual species of MLCL, DLCL, and CL in BTHS using high-resolution mass spectrometry, with implications for improved diagnostics and understanding of disease mechanisms.
Cell Samples:
Lymphoblastoid cell lines from 5 Barth syndrome (BTHS) patients and 5 controls were obtained from the Coriell Institute. Patient lines carried various TAZ mutations, including nonsense, missense, and full gene deletion.
Lipid Extraction:
Lipids were extracted from 1×10⁷ cells using a two-step chloroform/methanol protocol. An internal standard ((18:2)₄-CL d5) was added. Extracts were dried and stored at −80 °C.
LC-MS/MS Analysis:
Lipid profiling was performed using a Vanquish UHPLC coupled with an Orbitrap Fusion Tribrid ID-X mass spectrometer in negative ion mode. Separation was achieved on a C18 column with a 23-minute gradient run.
Data Analysis:
Lipid species were identified using LipidSearch 4.2 and quantified in Skyline by peak area normalization to the internal standard. Statistical differences were assessed using two-tailed Student's t-test with Benjamini-Hochberg correction.
Lipid Annotation via LC-MS/MS:
Comprehensive MS/MS analysis enabled structural identification of cardiolipin (CL), monolysocardiolipin (MLCL), and dilysocardiolipin (DLCL) species in lymphoblastoid cells. The composition of fatty acyl chains was determined based on precursor/product ion patterns, confirming dominant species such as (16:0)(18:0)(18:1)₂-CL.
Lipid Diversity and Abundance:
A wide range of CL, MLCL, and DLCL species with varying chain lengths and saturation was detected. Compared to controls, BTHS cells showed a ~50% reduction in total CL, while MLCL and DLCL increased ~20-fold and ~10-fold, respectively (adjusted P < 0.001). Species rich in oleic (18:1) and palmitoleic (16:1) acids were most abundant and significantly altered in BTHS.
Altered Lipid Profiles in BTHS:
Principal component analysis and clustering clearly separated BTHS and control groups. Heatmaps showed consistent upregulation of MLCL and DLCL, alongside downregulation of remodeled CL species. Notably, nascent CL (rich in palmitic/myristic acids) and MLCL precursors were increased, while mature CL was reduced. A standout example was (14:0)(16:0)(18:1)-MLCL, showing >350-fold elevation.
MLCL/CL and DLCL/CL Ratios:
Ratios of MLCL/CL and DLCL/CL were dramatically increased in BTHS cells. The total MLCL/CL ratio rose ~47-fold, and DLCL/CL ~22-fold (P < 0.001). Median ratios confirmed consistent increases (63-fold for MLCL/CL and 24-fold for DLCL/CL), aligning with disease-associated lipid remodeling defects.
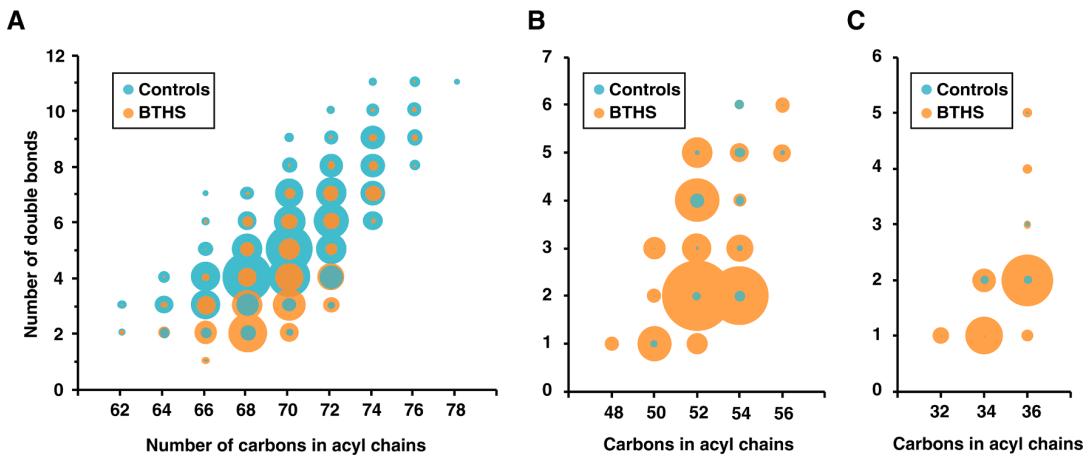 Diversity in cardiolipins, monolysocardiolipins and dilyosocardiolipins detected in the lymphoblastoid cells.
Diversity in cardiolipins, monolysocardiolipins and dilyosocardiolipins detected in the lymphoblastoid cells.
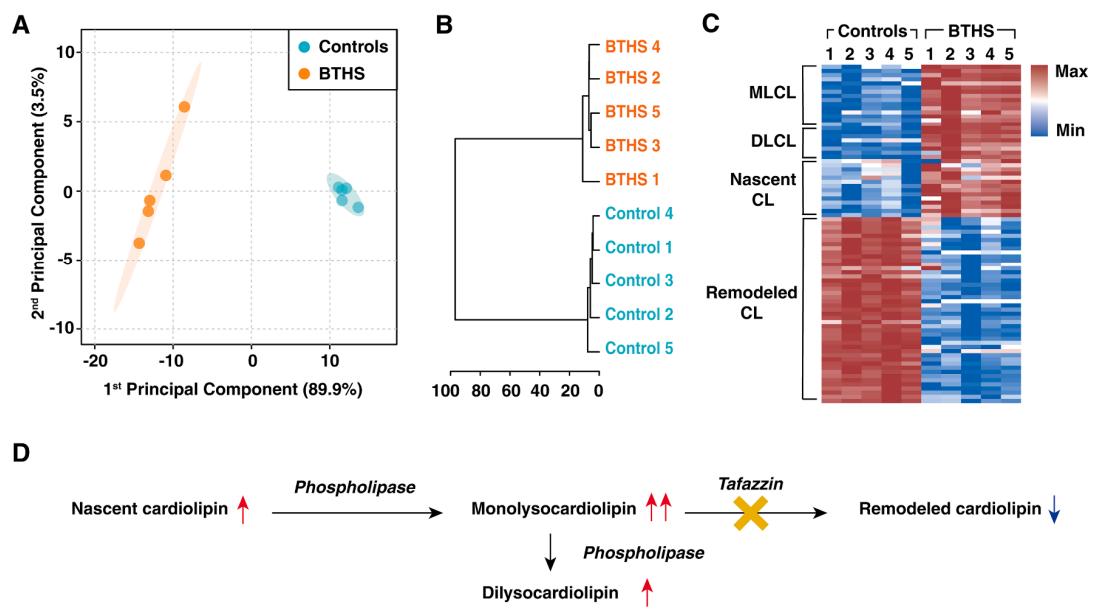 Distinct lipid profiling between 5 controls and 5 BTHS specimens.
Distinct lipid profiling between 5 controls and 5 BTHS specimens.
Reference
- Byeon, Seul Kee, et al. "High-resolution mass spectrometric analysis of cardiolipin profiles in Barth syndrome." Mitochondrion 60 (2021): 27-32. https://doi.org/10.1016/j.mito.2021.07.003
FAQ of Cardiolipin Analysis Service
How do you handle complex biological matrices (e.g., microbial or plant samples) to ensure accurate CL detection?
For microbial or plant samples, we employ bead-beating lysis combined with methanol-chloroform extraction under nitrogen atmosphere to minimize oxidation artifacts. For plants, mitochondrial enrichment protocols are applied to isolate CL-rich fractions. Samples are validated for purity via SDS-PAGE before analysis.
Can your service distinguish between CL species with similar molecular weights (e.g., CL(72:8) vs. CL(72:9))?
Yes. Our SCIEX Triple Quad™ 6500 LC-MS/MS System achieves a mass accuracy of<2 ppm, resolving CL isomers via optimized MRM transitions and retention time alignment. Post-processing software further validates species specificity.
What bioinformatics support is included for pathway analysis?
We provide Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway mapping, highlighting CL-related lipid remodeling pathways (e.g., CDP-DAG synthesis, PG/PA metabolism). Customized reports include fold-change heatmaps and PCA plots for multi-group comparisons.
Are CL oxidation products (e.g., oxidized CL (82:12)) detectable in your workflow?
Absolutely. Our method detects peroxidized CL species (e.g., hydroperoxy-CL) using precursor ion scanning for m/z 184→184 transitions. Oxidation levels are quantified against deuterated internal standards.
Can I analyze CL in extracellular vesicles (EVs) or mitochondria-depleted samples?
Yes. We optimize ultracentrifugation protocols to isolate EVs, followed by CL extraction with a sensitivity limit of 5 × 10⁶ EVs per sample. Mitochondria-depleted samples are validated via citrate synthase activity assays to confirm depletion.
How do you address batch-to-batch variability in longitudinal studies?
All samples from a study are processed in a single batch whenever possible. For multi-batch runs, isotopically labeled CL (64:0) is used as a universal internal standard, ensuring inter-batch CV<12%.
Is there a limit to the number of CL species analyzed per sample?
Our platform supports simultaneous quantification of 50+ CL species in a single injection. For larger panels (e.g., >100 species), we split analyses into parallel runs with overlapping QC samples to maintain data integrity.
Can you integrate CL data with other lipid classes (e.g., phosphatidylglycerol) for holistic lipidomics?
Yes. We offer add-on modules for co-analysis of PG, PA, and CDP-DAG, critical for studying CL biosynthesis. Data are normalized to total lipid content or mitochondrial protein concentration.
What is your policy for re-running failed samples?
If sample failure occurs due to technical errors (e.g., column contamination, MS calibration drift), we rerun at no extra cost. Client-provided samples with insufficient CL content are flagged during QC and require resubmission.
Learn about other Q&A.
Publications
Here are some publications in Lipidomics research from our clients:

- Annexin A2 modulates phospholipid membrane composition upstream of Arp2 to control angiogenic sprout initiation. 2023. https://doi.org/10.1096/fj.202201088R
- Glucosylceramide is essential for Heartland and Dabie bandavirus glycoprotein-induced membrane fusion. 2023. https://doi.org/10.1371/journal.ppat.1011232
- Loss of G0/G1 switch gene 2 (G0S2) promotes disease progression and drug resistance in chronic myeloid leukaemia (CML) by disrupting glycerophospholipid metabolism. 2022. https://doi.org/10.1002/ctm2.1146
- Experimental microbial dysbiosis does not promote disease progression in SIV-infected macaques. 2018. https://doi.org/10.1038/s41591-018-0132-5
- The olfactory receptor Olfr78 promotes differentiation of enterochromaffin cells in the mouse colon. 2024. https://doi.org/10.1038/s44319-023-00013-5
References
- Yuan, Zhi-Xin, et al. "Qualitative and quantitative lipidomic profiling of cardiolipin, oxidized cardiolipin, and monolysocardiolipin from human skeletal muscle by UPLC-Orbitrap-MSn." Analytica Chimica Acta 1350 (2025): 343825. https://doi.org/10.1016/j.aca.2025.343825
- Helmer, Patrick O., et al. "Mass spectrometric investigation of cardiolipins and their oxidation products after two-dimensional heart-cut liquid chromatography." Journal of Chromatography A 1619 (2020): 460918. https://doi.org/10.1016/j.chroma.2020.460918
- Bautista, Javier S., et al. "Advances in methods to analyse cardiolipin and their clinical applications." TrAC Trends in Analytical Chemistry 157 (2022): 116808. https://doi.org/10.1016/j.trac.2022.116808









