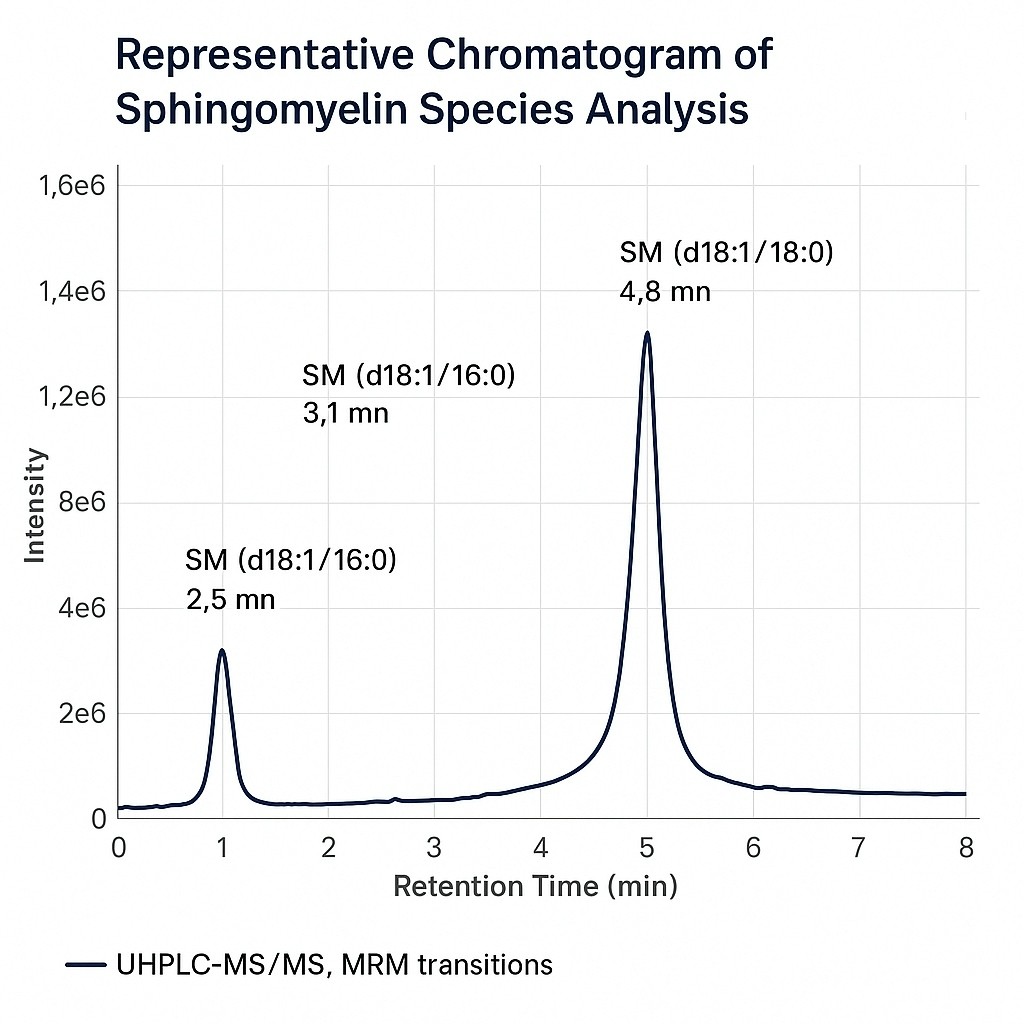
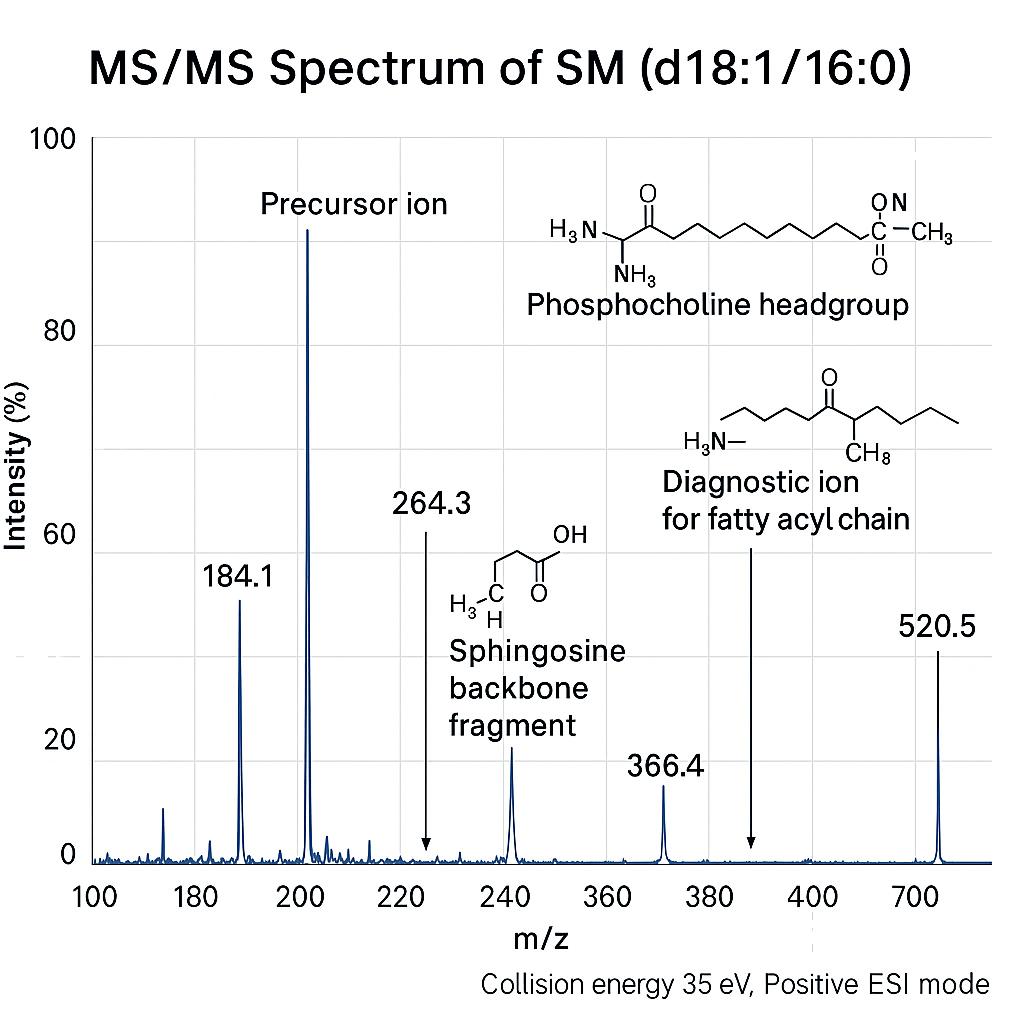
Sphingomyelin Profiling Service
Sphingomyelin is more than a structural lipid—it's a crucial player in cell signaling, lipid metabolism, and membrane dynamics. At Creative Proteomics, we empower researchers and industry partners with advanced sphingomyelin analysis, delivering clarity where lipid complexity obscures insights.
- Quantify over 50 sphingomyelin species with pg-level sensitivity
- Resolve isomers and structural variants using high-resolution MS/MS
- Analyze sphingomyelin alongside full sphingolipid panels
- Custom workflows for diverse matrices, from plasma to nutraceuticals
- Expert consultation and bioinformatics support for actionable data
Submit Your Request Now
×
What You'll Receive
- High-resolution chromatograms and mass spectra
- Comprehensive quantification tables with LOD and CV data
- Customizable reports for publication or further analysis
- Scientific interpretation and follow-up support
- What We Provide
- Technology Platform
- Sample Requirement
- Demo
- Case
- FAQ
What is Sphingomyelin?
Sphingomyelin is a complex sphingolipid composed of a ceramide backbone linked to a phosphocholine or phosphoethanolamine headgroup. As a major structural lipid in eukaryotic membranes, it plays critical roles in maintaining membrane architecture, lipid raft formation, and regulating membrane fluidity.
Beyond structural functions, sphingomyelin is a bioactive lipid involved in key cellular processes, including:
- Signal transduction
- Regulation of apoptosis
- Cell differentiation
- Lipid metabolism cross-talk
Its catabolism generates ceramide and other signaling molecules pivotal for cellular homeostasis. Disruptions in sphingomyelin metabolism are increasingly recognized in diverse biological contexts, underscoring the need for precise sphingomyelin profiling in both research and industrial applications.
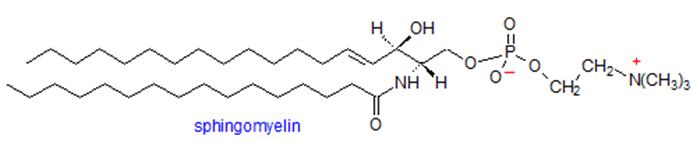 Structure of sphingomyelin
Structure of sphingomyelin
Our Specialized Sphingomyelin Profiling Services
- Quantitative Profiling of Sphingomyelin Species: Accurate measurement of individual sphingomyelin molecular species across various sample types, including plasma, tissues, and cultured cells.
- Qualitative Identification and Characterization: Detailed structural analysis to differentiate sphingomyelin variants by acyl chain length, saturation, and headgroup modifications.
- Integrated Sphingolipid Panel Analysis: Concurrent detection of sphingomyelin alongside related sphingolipids such as ceramides and sphingosine-1-phosphate, providing a holistic view of sphingolipid metabolism.
- Customized Sample Preparation Protocols: Optimized lipid extraction methods tailored to preserve sphingomyelin integrity and maximize recovery from specific biological matrices.
- Targeted and Untargeted Lipidomics Approaches: Flexible analytical strategies that cater to hypothesis-driven research or broad lipid discovery projects.
- Data Processing and Bioinformatics Support: Comprehensive data analysis including lipid identification, quantification, and pathway interpretation to facilitate actionable scientific insights.
- Project-Specific Method Development: Development and validation of specialized assays for unique sample types or novel sphingomyelin species upon request.
List of Detected Sphingomyelin and Related Metabolites
| Lipid Class | Representative Analytes |
|---|---|
| Sphingomyelin (SM) | SM (d18:1/14:0), SM (d18:1/16:0), SM (d18:1/18:0), SM (d18:1/18:1), SM (d18:1/20:0), SM (d18:1/22:0), SM (d18:1/24:0), SM (d18:1/24:1), SM (d18:1/26:0), SM (d18:1/26:1) |
| Ceramide (Cer) | Cer (d18:1/14:0), Cer (d18:1/16:0), Cer (d18:1/18:0), Cer (d18:1/20:0), Cer (d18:1/22:0), Cer (d18:1/24:0), Cer (d18:1/24:1), Cer (d18:1/26:0) |
| Dihydroceramide (dhCer) | dhCer (d18:0/16:0), dhCer (d18:0/18:0), dhCer (d18:0/24:0) |
| Hexosylceramide (HexCer) | HexCer (d18:1/16:0), HexCer (d18:1/24:1), HexCer (d18:1/24:0) |
| Lactosylceramide (LacCer) | LacCer (d18:1/16:0), LacCer (d18:1/24:1) |
| Sphingosine (So) | d18:1 Sphingosine |
| Sphinganine (Sa) | d18:0 Sphinganine |
| Lysosphingomyelin (LSM) | LSM (d18:1), LSM (d18:0) |
| Sphingosine-1-Phosphate (S1P) | d18:1 S1P |
| Dihydrosphingosine-1-Phosphate (dhS1P) | d18:0 dhS1P |
Note: Custom analyte panels and additional species are available upon request to suit specific research needs
Methods and Instrumentation for Sphingomyelin Analysis
Agilent 6495C Triple Quadrupole LC-MS/MS
- Ultra-high sensitivity with LOD<10 pg on-column for sphingomyelin species.
- Operates in MRM mode for precise quantification across 4–5 orders of magnitude.
- Positive ESI mode ensures reliable detection of phosphocholine headgroups.
 Agilent 6495C Triple quadrupole (Figure from Agilent)
Agilent 6495C Triple quadrupole (Figure from Agilent)
Agilent 7890B-5977A GC-MS
- Ideal for analyzing volatile lipid derivatives or fatty acid fragments.
- Electron ionization (EI) provides reproducible spectra for lipid structure confirmation.
- Ensures high mass accuracy and stability for complex lipid profiling.
 Agilent 7890B-5977A (Figure from Agilent)
Agilent 7890B-5977A (Figure from Agilent)
Agilent 1260 Infinity II HPLC
- High-resolution separation of sphingomyelin species with sub-2 μm particle columns.
- Supports both reverse-phase and normal-phase methods.
- Handles pressures up to 600 bar for sharp peak resolution.
 Agilent 1260 Infinity II HPLC (Figure from Agilent)
Agilent 1260 Infinity II HPLC (Figure from Agilent)
Analytical Workflow: Your Path to Reliable Sphingomyelin Data
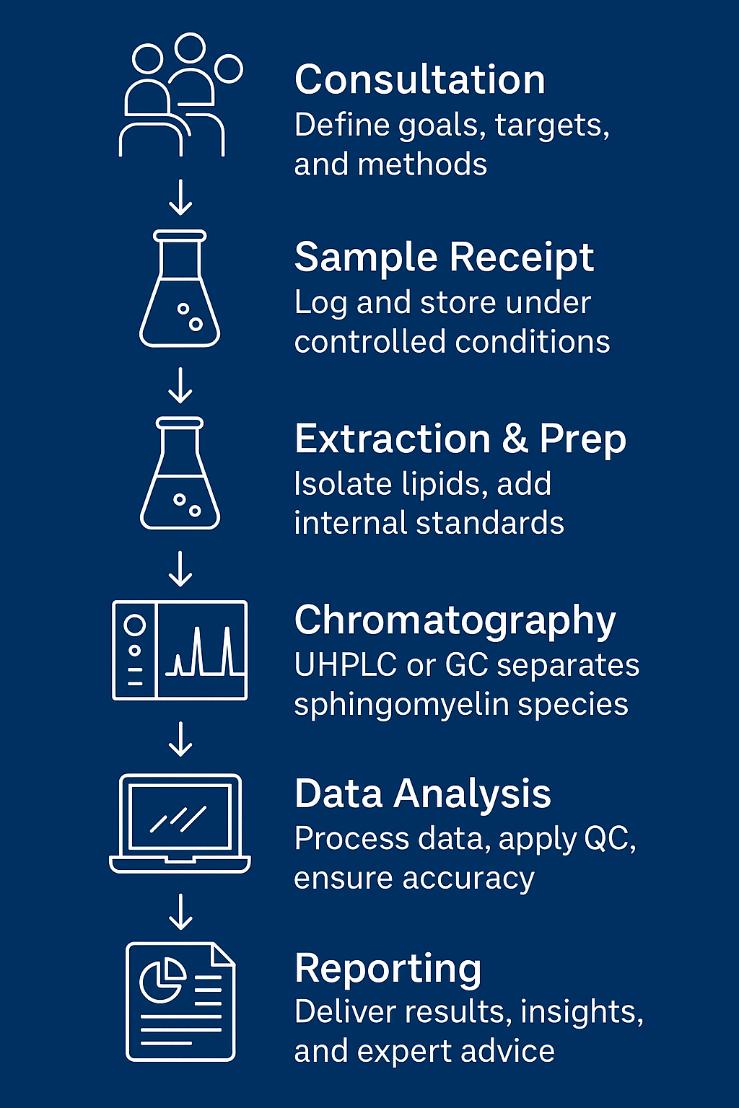
Sample Requirements for Sphingomyelin Analysis Service
| Sample Type | Recommended Amount | Notes |
|---|---|---|
| Serum / Plasma | ≥ 50 μL | Preferably stored at -80°C |
| Cell Pellets | ≥ 1 × 10⁶ cells | Washed to remove media before freezing |
| Tissue Samples | ≥ 10 mg | Fresh-frozen, avoid prolonged storage |
| Cerebrospinal Fluid (CSF) | ≥ 100 μL | Minimum volume for low-abundance detection |
| Food / Nutraceuticals | ≥ 50 mg | Powdered or homogenized recommended |
| Lipid Extracts | ≥ 10 μg total lipids | Indicate extraction solvents used |
| Urine | ≥ 500 μL | Optional, for exploratory studies |
| Culture Media / Supernatant | ≥ 500 μL | For cell-based sphingolipid studies |
Demo Results
Sphingomyelin Analysis Service Case Study

Title: LC-MS Based Sphingolipidomic Study on A2780 Human Ovarian Cancer Cell Line and its Taxol-resistant Strain
Journal: Scientific Reports
Published: 2016
- Study Summary
- Background
- Methods
- Results
- Reference
This study employed a liquid chromatography-mass spectrometry (LC-MS) based sphingolipidomic approach to investigate the sphingolipid metabolism in both taxol-sensitive and taxol-resistant A2780 human ovarian cancer cell lines. The research identified 102 sphingolipid species, 67 of which were newly reported for the A2780 cell line. The study highlighted significant changes in sphingolipid profiles, especially in sphingomyelin and ceramide subclasses, which were linked to taxol resistance. The findings suggest that dysregulation in sphingolipid metabolism, particularly the depletion of ceramides and the increase in sphingomyelins, may be a crucial mechanism for taxol resistance in ovarian cancer cells.
Ovarian cancer remains one of the deadliest cancers, with resistance to chemotherapeutic drugs, particularly taxol, posing significant challenges in treatment. Taxol resistance in ovarian cancer is associated with various mechanisms, including alterations in drug transport, apoptosis evasion, and metabolic changes. Recent studies have pointed to sphingolipids—important components in cell membrane structure and signaling—as being involved in the regulation of drug resistance. However, comprehensive sphingolipidomic studies on taxol-resistant ovarian cancer cells had been limited, making this study crucial for understanding how sphingolipid metabolism influences chemoresistance.
Sphingolipid Extraction
A2780 and A2780T cells were cultured and sphingomyelins were extracted using methanol, chloroform, and an internal standard cocktail.
LC-MS Conditions
Sphingomyelins were separated using an Agilent 1290 UHPLC system with a C18 column. A Q-TOF mass spectrometer was used for qualitative analysis, and a triple-quadrupole MS (QQQ) was employed for quantitative analysis using MRM.
Data Analysis
Sphingomyelin profiles were analyzed with PLS-DA to identify significant differences between A2780 and A2780T cells.
![]() Creative Proteomics offers precise sphingomyelin analysis services to advance cancer research.
Creative Proteomics offers precise sphingomyelin analysis services to advance cancer research.
We provide:
- Accurate sphingomyelin quantification in cancer cell lines
- High-resolution LC-MS/MS platforms for profiling sphingolipids
- Customized solutions to explore sphingolipid metabolism in drug-resistant models like A2780T
The study successfully identified 102 sphingolipid species in both A2780 and its taxol-resistant strain, A2780T. Notably, the analysis revealed significant differences between the two cell lines, including a decrease in ceramide levels and an increase in sphingomyelin content in the taxol-resistant cells. Among the identified sphingolipids, 52 species showed significant alterations in A2780T cells compared to the sensitive strain. The study concluded that these metabolic changes in sphingolipid profiles could play a critical role in the development of taxol resistance. Key findings included:
- Increased sphingomyelins in A2780T cells, particularly with a C16 sphingoid base.
- Decreased ceramides, hexosylceramides, and lactosylceramides in A2780T cells.
- A potential alteration in sphingolipid metabolism pathways, particularly related to the sphingomyelin/ceramide pathway, which could contribute to the evasion of ceramide-induced apoptosis in resistant cells.
These findings provide valuable insights into the mechanisms of taxol resistance, with implications for the development of targeted therapies based on sphingolipid metabolism.
Reference
- Huang, Hao, et al. "LC-MS based sphingolipidomic study on A2780 human ovarian cancer cell line and its taxol-resistant strain." Scientific reports 6.1 (2016): 34684. https://doi.org/10.1038/srep34684
FAQ of Sphingomyelin Analysis Service
Can Creative Proteomics distinguish between positional isomers of sphingomyelin species?
Yes. Using high-resolution MS/MS and optimized chromatographic conditions, we can differentiate sphingomyelin isomers based on acyl chain position and unsaturation patterns.
What is the typical quantitation accuracy for sphingomyelin analysis?
For most sphingomyelin species, quantitation accuracy is within ±10% RSD, supported by calibration curves using isotope-labeled standards.
Are matrix effects a concern in sphingomyelin analysis, and how are they addressed?
Matrix effects are evaluated through spike-recovery experiments. We apply matrix-matched calibration whenever feasible to ensure reliable quantification in complex samples.
Can you integrate sphingomyelin data with other omics datasets?
Yes. We provide data in formats compatible with multi-omics integration and can assist with data normalization for cross-platform analyses.
Is there a limit to how many sphingomyelin species can be analyzed in a single run?
Our targeted methods typically accommodate 50–100 sphingomyelin species. For untargeted workflows, we can profile over 200 sphingolipid species, depending on sample complexity and instrumentation.
Do you analyze both saturated and unsaturated sphingomyelin species?
Absolutely. Our methods resolve both saturated and unsaturated forms, including mono- and polyunsaturated species up to C26 or longer chain lengths.
How do you ensure the stability of sphingomyelin during sample processing?
We use low-temperature extractions, antioxidants where necessary, and minimize exposure to light and oxygen to preserve lipid integrity throughout preparation.
How low can you detect sphingomyelin in biological fluids like CSF?
Detection limits in CSF are typically in the low pg/mL range, depending on the specific species and matrix background.
Learn about other Q&A about proteomics technology.
Publications
Here are some of lipidomics-related papers published by our clients:

- White matter lipid alterations during aging in the rhesus monkey brain. 2024. https://doi.org/10.1007/s11357-024-01353-3
- Characterization of Dnajc12 knockout mice, a model of hypodopaminergia. 2024. https://doi.org/10.1101/2024.07.06.602343
- Evidence for phosphate-dependent control of symbiont cell division in the model anemone Exaiptasia diaphana. 2024. https://doi.org/10.1128/mbio.01059-24
- The olfactory receptor Olfr78 promotes differentiation of enterochromaffin cells in the mouse colon. 2024. https://doi.org/10.1038/s44319-023-00013-5
- Annexin A2 modulates phospholipid membrane composition upstream of Arp2 to control angiogenic sprout initiation. 2023. https://doi.org/10.1096/fj.202201088R
- Lipid Membrane Engineering for Biotechnology. 2023. https://doi.org/10.48780/publications.aston.ac.uk.00046663