Hexosylceramide Analysis Service
Creative Proteomics delivers targeted LC-MS/MS for HexCer (GlcCer/GalCer) with true isomer separation, isotope-standard quantitation, and transparent QC so you can trust every comparison and defend every conclusion.
- Isomer-Resolved Quantification – Differentiate GlcCer vs GalCer via HILIC-LC separation
- Trace-Level Sensitivity – Matrix-optimized extraction and isotope dilution
- QC-Integrated Data – Pooled sample tracking, calibration curves, precision stats
- Validated Instrument Platforms – Orbitrap & Triple Quad systems with locked parameters
- Broad Chain-Length Coverage – C14–C26, saturated and unsaturated HexCer species
Submit Your Request Now
×
Deliverables
- Raw Data: LC-MS/MS vendor files with open-format conversions
- Quant Tables: GlcCer/GalCer concentrations, chain-length resolved
- QC Summary: Calibration, precision, drift monitoring
- Statistical Plots: Heatmaps, PCA, fold-change charts
- What We Provide
- Advantages
- Technology Platform
- Sample Requirement
- Demo
- FAQs
What Is Hexosylceramide (HexCer)?
Hexosylceramides are glycosphingolipids comprising a ceramide backbone glycosylated with a single hexose (glucose or galactose). As central nodes in sphingolipid metabolism, HexCer participate in membrane organization, trafficking, and signaling. Their chain-length and unsaturation patterns shift with cellular state, making HexCer a sensitive readout for pathway modulation, lipid remodeling, and stress responses in discovery research, mechanism studies, and bioprocess monitoring.
What Problems We Solve for Our Clients
Struggling with low-abundance detection in complex biological matrices?
We implement matrix-optimized extraction protocols, internal standards, and ion suppression mitigation strategies to ensure reliable detection and quantitation of trace-level HexCer species.
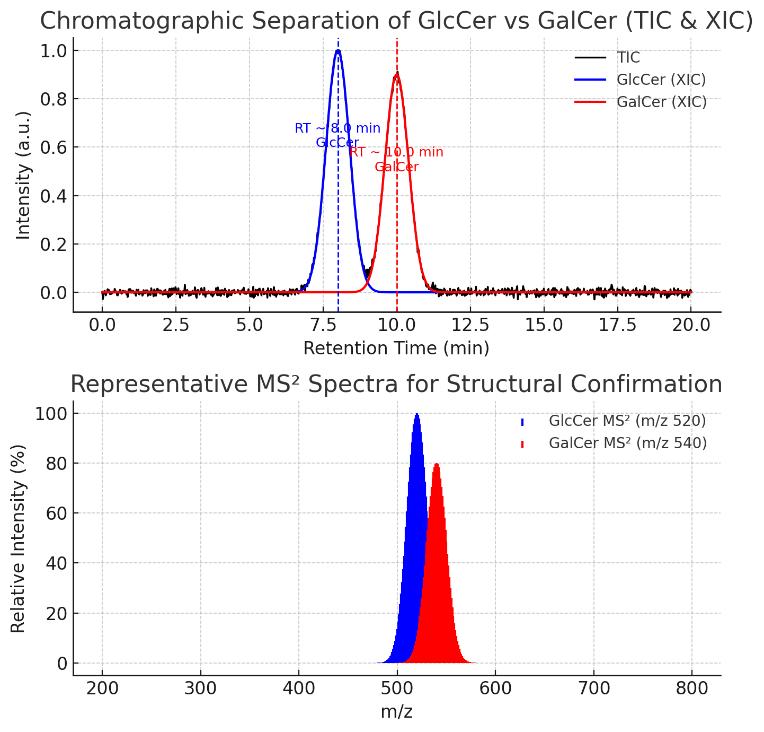
Need to resolve isobaric/isomeric species like GlcCer vs. GalCer?
Our workflow includes HILIC-based chromatographic separation and diagnostic fragment monitoring to distinguish structurally similar HexCer subclasses with high specificity.

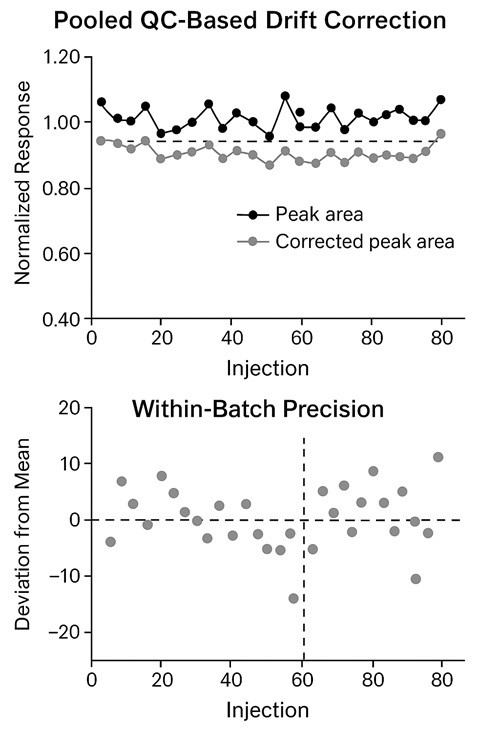
Lacking consistent data across large cohorts or multi-batch studies?
With rigorous QC strategies—including pooled sample tracking, RT-locking, and normalized quantification—we minimize technical variability and support longitudinal comparability.
Facing limitations with standard lipidomics platforms for pathway-level insights?
Our targeted method covers a wide range of sphingoid base and N-acyl chain combinations (C14–C26), enabling high-resolution profiling for lipid metabolism, cellular stress, and membrane remodeling studies.
Creative Proteomics's Hexosylceramide Analysis Solutions
- Targeted HexCer (GlcCer, GalCer) panel quantification by LC-MRM
- Expanded long-chain and very-long-chain HexCer coverage (C14–C26, saturated/unsaturated)
- Absolute quantitation with isotopically-labeled standards where available; otherwise high-rigor relative quantitation
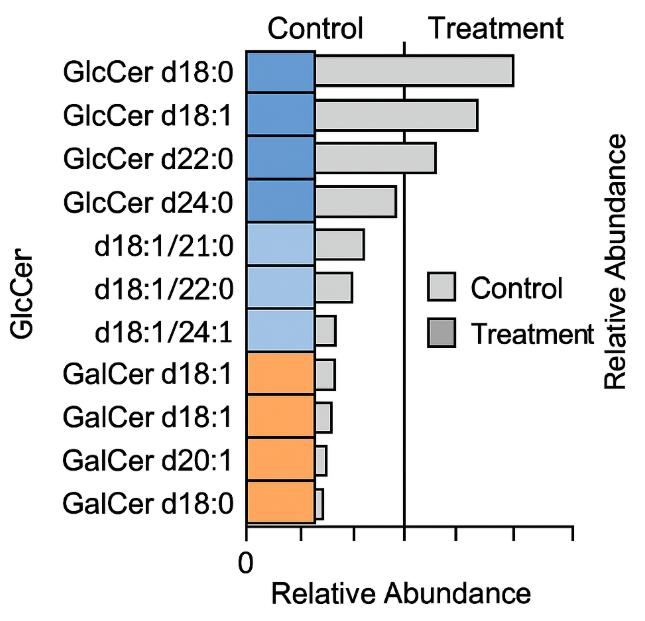
- Subclass-resolved reporting (Hex1Cer: GlcCer vs GalCer) with typical sphingoid bases (d18:1, d18:0, d20:1)
- Method transfer / cross-site alignment (calibrators, RT locks, ion ratio checks)
- Biostat add-ons: group comparisons, PCA, heatmaps, pathway overlays
Advantages of Our Hexosylceramide Analysis Services
- Mass accuracy: ≤ 3 ppm (Orbitrap) for ID confidence
- Dynamic range: ≥ 10³–10⁴ (calibration dependent)
- Intra-batch precision: CV ≤ 10% for QCs; inter-batch CV typically ≤ 15%
- Recovery: 85–115% (matrix-matched) with isotope dilution where standards exist
- Carryover: < 0.1% with back-flush and needle-wash controls
- Peak capacity: ≥ 150 features per 20-min gradient (HexCer panel + sentinels)
- RT stability: pooled-QC RT RSD ≤ 0.5% with column-temp control and RT lock
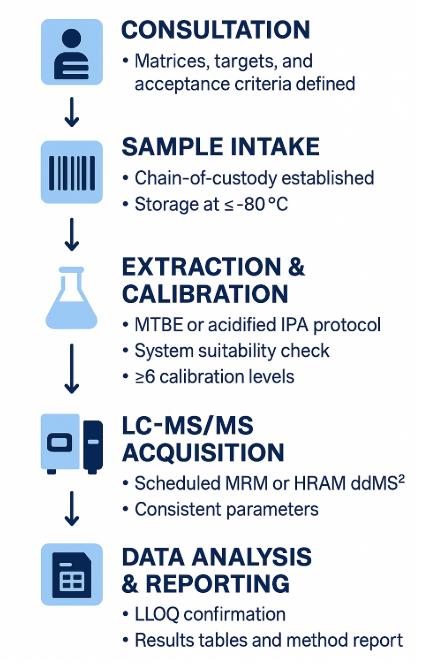
Workflow for Hexosylceramide Analysis Service
1. Consultation & panel lock-in — matrices, targets, reporting units, and acceptance criteria are defined.
2. Sample intake & barcoding — chain-of-custody established; storage at ≤ −80 °C.
3. Extraction with internal standards — MTBE-based or acidified IPA protocol; recovery sentinels included.
4. System suitability — mass-cal, RT lock mix, blanks, and carryover check.
5. Calibration & QCs — ≥6 levels (when absolute quant) plus pooled-QC every 10 samples for drift tracking.
6. LC-MS/MS acquisition — scheduled MRM or HRAM ddMS² with consistent parameters.
7. Data processing — peak integration, ion-ratio/RT verification, blank subtraction, and batch normalization if pre-approved.
8. QA review & reporting — precision/accuracy tables, LLOQ confirmation, and flagged-analyte notes.
9. Delivery — results tables, raw/processed files, and method report for reproducibility.
Technology Platform for Hexosylceramide Analysis Service
Chromatography
UHPLC systems: Thermo Vanquish or Waters ACQUITY UPLC
Columns (typical)
- HILIC: ACQUITY BEH Amide, 1.7 µm, 2.1 × 100 mm (HexCer subclass separation; GlcCer vs GalCer)
- RP backup: C18, 1.7 µm, 2.1 × 100 mm (acyl-chain resolution)
Mobile phases
- A: ACN:H₂O with 10 mM ammonium formate + 0.1% formic acid
- B: IPA:ACN with 10 mM ammonium formate + 0.1% formic acid
Gradient: 20–22 min typical; flow 0.3–0.4 mL/min; column temp 40 °C; injection 2–5 µL
Mass Spectrometry Platforms
High-resolution: Thermo Q Exactive™/Exploris™ (Orbitrap)
- Polarity: ESI+ (primary), optional ESI− confirmation
- Resolution: 70k–120k @ m/z 200 (full scan), 17.5k–35k (ddMS²)
- AGC: 1e6 (full), 2e5 (MS²); max IT tuned for ≥10 points/peak
Triple Quadrupole: SCIEX 6500+/7500
- Acquisition: scheduled MRM; cycle time 1.5–2.0 s; dwell 5–20 ms
- CE: optimized per backbone (e.g., 25–45 eV for HexCer d18:1)
- Transitions: [M+H] → 264.3 (d18:1), 266.3 (d18:0), 292.3 (d20:1), with qualifier ions for ion-ratio checks
Quantitation & ID Rules
Primary: isotope-dilution where available; otherwise matrix-matched external calibration
ID confidence: RT match ±0.15 min to standards, MS/MS fragment match, mass accuracy ≤3 ppm (HRAM) or ion-ratio within tolerance (MRM)
Data systems: TraceFinder™, LipidSearch™, Skyline, custom R scripts for QC charts
Sample Requirements for Hexosylceramide Analysis Service
| Matrix | Minimum Amount | Container | Stabilization & Storage | Notes |
|---|---|---|---|---|
| Plasma/Serum | ≥ 50 µL | Low-bind microtube | Freeze at ≤ −80 °C; avoid repeated freeze-thaw | Record anticoagulant; EDTA preferred |
| Whole Blood | ≥ 200 µL | Low-bind tube | Chill promptly; separate plasma if possible | Hemolysis may impact lipid background |
| Cell Pellet | ≥ 1 × 10⁶ cells or ≥ 20 µL packed | Cryovial | Wash PBS (no serum), snap-freeze | Provide cell count and buffer details |
| Tissue | 20–30 mg | Cryovial/foil | Snap-freeze; store ≤ −80 °C | Homogenize on dry ice if pre-processed |
| CSF/Urine | ≥ 200 µL | Low-bind tube | Store ≤ −80 °C | Creatinine or protein normalization optional |
| Culture Medium | ≥ 500 µL | Low-bind tube | Store ≤ −80 °C | Note serum content and timepoint |
| Lipid Extract | ≥ 50 µL | Amber vial | Dry under N₂; store ≤ −20 °C, dark | Provide extraction recipe for compatibility |
Demo Results
FAQ of Hexosylceramide Analysis Service
What is the minimal detectable quantity (LOD/LOQ) for low-abundance HexCer species?
LOQ typically reaches low nanomolar or sub-nanomole levels depending on the species, matrix, and ionization efficiency. When stable isotope-labeled internal standards are used, LOD and LOQ are empirically determined and reported per analyte in the validation data.
Can chromatographic retention time alone differentiate GlcCer and GalCer?
Yes, under HILIC separation with BEH amide columns, GlcCer and GalCer isomers exhibit distinct retention profiles despite identical m/z. Retention time windows are locked using chemical standards and QC mixtures, and deviations beyond ±0.15 min are flagged during data processing.
Why do I see elevated HexCer levels in disease models or stressed cells?
Elevated HexCer species can reflect disrupted sphingolipid homeostasis, impaired lysosomal degradation, or enhanced salvage pathway activity. In disease contexts, such changes may indicate compensatory mechanisms, mitochondrial stress, or inflammatory signaling. Profiling HexCer dynamics can help map these biochemical responses.
How does HexCer profiling enhance lipidomics studies beyond total ceramide measurement?
While total ceramide levels capture the endpoint of sphingolipid catabolism, HexCer profiling adds a layer of resolution at the glycosylation interface. Subclass-resolved HexCer data can reveal upstream metabolic shifts or substrate accumulation that would be missed in bulk ceramide quantitation alone.
Are Hexosylceramides relevant in non-mammalian or engineered systems?
Yes. HexCer species have been identified in microbial systems, plants, and synthetic biology models where sphingolipid glycosylation is engineered or modulated. Their detection supports host-pathogen interaction studies, microbial virulence profiling, and biomanufacturing lipid quality control.
How is the specificity of HexCer analysis ensured in complex samples?
Specificity comes from combining chromatographic resolution, accurate mass (or MRM transitions), diagnostic MS/MS fragmentation, and retention time locking. When isotope-labeled internal standards are included, quantitation accuracy and ID confidence improve further.
Can I integrate HexCer results with pathway enrichment or network analysis?
Yes. HexCer data can be mapped onto sphingolipid metabolic pathways, annotated with chain-length and saturation attributes, and integrated into systems biology frameworks such as lipid-centric PCA, clustering, or flux analysis. This adds mechanistic depth to omics datasets.
How does this analysis handle low-abundance or minor HexCer species?
Low-abundance HexCer are addressed via signal-enhancing extraction protocols, background suppression, and scheduled acquisition windows. In some cases, quantitation is semi-quantitative (relative), but still reproducible and valid for comparative or mechanistic studies.
Do I need to know my target species before requesting analysis?
Not necessarily. The analysis can begin with a validated HexCer panel spanning typical chain lengths (C14–C26), and expand based on exploratory results. You may define targets in advance, or use an initial screen to inform subsequent focused quantitation.
Can I compare my HexCer results across studies or timepoints?
Yes, provided pooled QCs, consistent acquisition parameters, and locked retention times are used. Our output is designed to support longitudinal, multi-batch, or multi-site comparisons, minimizing drift and ensuring reproducibility across experimental conditions.
Learn about other Q&A about proteomics technology.
Publications
Here are some of the lipidomics-related papers published by our clients:

- White matter lipid alterations during aging in the rhesus monkey brain. 2024.
- Characterization of Dnajc12 knockout mice, a model of hypodopaminergia. 2024.
- Annexin A2 modulates phospholipid membrane composition upstream of Arp2 to control angiogenic sprout initiation. 2023.
- Lipid Membrane Engineering for Biotechnology (Doctoral dissertation, Aston University). 2023.
- Summative and ultimate analysis of live leaves from southern US forest plants for use in fire modeling. 2020.