High-Resolution Protein Acylation Profiling via LC-MS/MS
Our Acylation Quantitative Proteomics Service enables researchers to precisely measure and compare acylation patterns across biological samples under different physiological or experimental conditions. By mapping changes in site-specific acylation, you can:
- Identify dynamic protein regulation under stress, drug treatment, or disease states
- Uncover mechanistic links between acylation and cellular pathways
- Discover potential biomarkers or therapeutic targets for further research
Whether you're investigating epigenetic control mechanisms or exploring novel metabolic regulators, our service provides high-resolution, quantitative acylation profiles using advanced LC-MS/MS technology and tailored enrichment strategies.
Submit Your Request Now
×
Get insights into acylation-driven regulation with our high-throughput proteomics platform.
- What is
- Service Details
- FAQ
- Case Study
- Publication
What is Protein Acylation
Protein acylation is gaining traction as a critical regulator in modern proteomics. This post-translational modification (PTM) involves the transfer of acyl groups—like acetyl, succinyl, or crotonyl—from acyl-CoA metabolites directly onto proteins. These modifications influence key cellular functions including gene regulation, energy metabolism, protein transport, and protein–protein interactions.
While acetylation remains the most well-characterized acylation event, newer forms—such as propionylation, malonylation, glutarylation, and crotonylation—are now expanding our view of cellular dynamics and proteome complexity.
Why Protein Acylation Matters
Changes in acylation patterns often signal shifts in physiological or pathological states. By comparing acylation levels under different conditions, researchers can:
- Map signaling pathways and epigenetic control mechanisms
- Identify biomarkers for disease detection
- Discover novel drug targets that impact therapeutic development
In short, acylation profiling offers a powerful lens for understanding how cells respond to internal and external stimuli.
Our Protein Acylation Analysis Service
At Creative Proteomics, we've developed a full-service solution for acylated proteomics profiling. Powered by Thermo Fisher's Orbitrap Fusion Lumos and integrated with high-precision nanoLC-MS/MS, our platform delivers high-resolution mapping of acylated proteins across diverse sample types.
Whether you're investigating chromatin dynamics, metabolic regulation, or PTM crosstalk, our service provides accurate, actionable data to support your research goals.
Our Protein Acylation Profiling Packages
To support a wide spectrum of research applications—from epigenetics and energy metabolism to signal transduction and membrane biology—our protein acylation analysis service offers modular profiling packages across three major acylation subtypes: N-terminal acylation, lysine side-chain acylation, and S-acylation. Each category is supported by specialized protocols and targeted LC-MS/MS workflows.
Service Packages
- N-terminal Acylation
- Lysine Acylation
- S-acylation
N-terminal Acylation
A prevalent, irreversible modification that stabilizes proteins and affects degradation pathways.
N-myristoylation
A lipid-based modification where myristic acid is added to the N-terminus, often regulating protein-membrane association and intracellular signaling.
Lysine Acylation
We offer targeted detection of lysine acylation subtypes, each linked to distinct biological functions and research domains:
Analyze epigenetic regulation and chromatin remodeling.
Analyze protein regulation related to fatty acid metabolism and short-chain acyl-CoA derivatives.
Malonylation
Examine mitochondrial protein function and oxidative stress modulation.
Glutarylation
Investigate lysine metabolism, neurodevelopmental processes, and metabolic pathway regulation.
Explore protein roles in the TCA cycle and energy production.
→ Supports studies in metabolic PTMs and mitochondrial biology.
Map chromatin dynamics and transcriptional activation in epigenetic research.
2-Hydroxyisobutyrylation, butyrylation, hexanoylation, and others
Available on request for custom acylation profiling projects involving less-characterized PTMs.
S-acylation
A reversible thioester linkage of palmitic acid to cysteine residues, crucial for membrane anchoring and signal transduction.
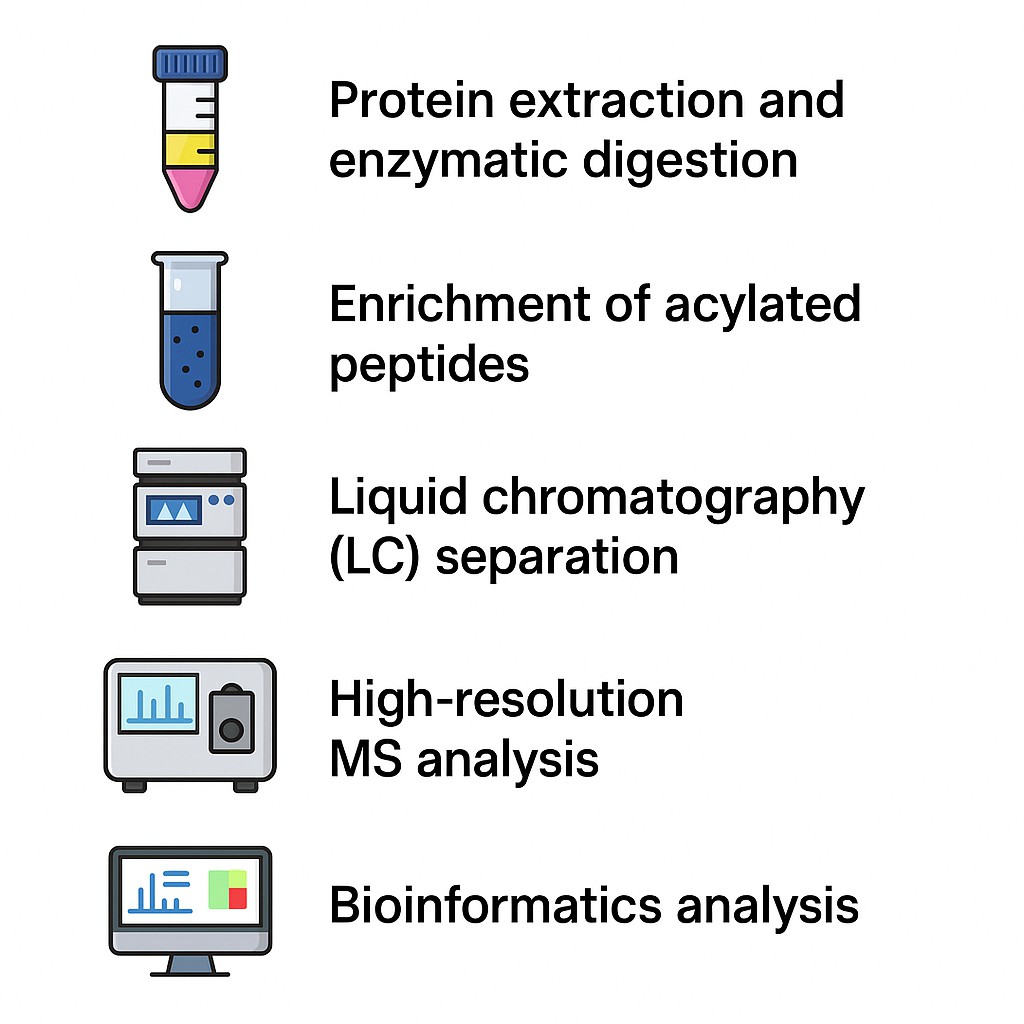
Our Acylation Proteomics Workflow
We manage the entire workflow, from sample prep to data interpretation:
- Protein extraction and enzymatic digestion
- Enrichment of acylated peptides using high-affinity antibodies or proprietary kits
- Liquid chromatography (LC) separation to reduce sample complexity
- High-resolution MS analysis for peptide identification and quantification
- Bioinformatics analysis, including acylation site annotation, functional classification, and enrichment studies. You'll receive a complete dataset detailing acylated modification sites and abundance levels, along with expert-curated insights into functional implications.
High-Confidence Results You Can Trust
By integrating antibody-based enrichment with cutting-edge mass spectrometry, our method ensures high specificity and sensitivity. This allows researchers to:
- Detect even low-abundance modifications
- Quantify site-specific acylation events
- Explore PTM interplay in complex biological systems
Our workflow is optimized for a wide range of organisms, provided a well-annotated genome or protein sequence database is available.
Why Choose Creative Proteomics for Acylation Profiling?
Our acylation quantitative proteomics platform combines analytical depth, technical reproducibility, and biological interpretability. Here's what you can expect when partnering with us:
- Site-Specific Acylation Mapping with >95% Confidence
We deliver high-resolution identification of acylation sites using optimized MS workflows and validated software pipelines, ensuring confident localization even for low-abundance targets.
- High-Specificity Enrichment of Modified Peptides
Our workflows incorporate validated antibodies and commercially optimized resin kits for acylation-specific enrichment. This targeted approach significantly increases sensitivity and reduces background noise, even in complex proteomes.
- Broad Coverage of Acylation Subtypes
In addition to standard lysine acetylation, we support comprehensive profiling of propionylation, succinylation, malonylation, crotonylation, palmitoylation, and other acyl-CoA–linked modifications.
- Scalable LC-MS/MS Detection of Thousands of Sites
Our high-throughput mass spectrometry platforms enable quantification of thousands of acylation sites per run, making it suitable for both discovery-phase and comparative studies.
- Flexible Quantification Options
We support multiple labeling strategies—including TMT, SILAC, and label-free quantification—to accommodate both relative and absolute comparisons across experimental conditions.
- Compatibility with Diverse Sample Types
Whether working with cell lines, tissues, biofluids, or model organisms, our protocols are optimized to preserve modification integrity and maximize proteome coverage.
- Biological Interpretation with Integrated Bioinformatics
We include pathway analysis, protein domain mapping, and functional annotation to help translate acylation data into mechanistic insights.
- High-specificity enrichment of modified peptides using validated antibodies and resin kits
- Scalable LC-MS/MS detection of thousands of acylation sites in a single run
- Compatibility with TMT, SILAC, and label-free quantification methods for direct comparison across conditions
Platform

Orbitrap Fusion Lumos

nanoLC-MS/MS
Delivery
You will receive a complete analysis package including:
- Raw MS data files
- Identified acylated peptides with site-level annotation
- Quantitative intensity tables
- Summary reports of top pathways and modified proteins
- Optional: annotated spectra, GO enrichment, and protein interaction maps
Sample Requirements
| Sample Type | Minimum Amount Required | Notes |
|---|---|---|
| Fresh Animal Tissue | ≥ 600 mg | Snap-freeze in liquid nitrogen and store at −80°C |
| Fresh Plant Tissue | ≥ 6 g | Remove fibrous materials and clean before freezing |
| Cell Culture | ≥ 1 × 10⁷ cells/tube × 3 tubes | Adherent/suspension cells both acceptable; avoid serum contamination |
| Fungi and Bacteria | ≥ 600 mg | Pellet and freeze-dry for stability if long-term storage needed |
| Serum or Plasma | 450 μL × 4 tubes | Store in polypropylene tubes; avoid repeated freeze-thaw |
| Protein Solution | 5–10 mg total protein | Concentration ≥1 μg/μL preferred; provide buffer composition |
| Urine | 15 mL × 4 tubes | Centrifuge at 1000 × g for 5 min; discard pellet |
| Other Body Fluids | >15 mL | Includes saliva, amniotic fluid, CSF, etc.; please clarify matrix |
Frequently Asked Questions (FAQ)
Can you analyze low-abundance acylation sites in complex tissues like brain or liver?
Yes. Our workflow integrates high-affinity acyl-peptide enrichment (antibody- or resin-based) with deep LC-MS/MS coverage, enabling detection of site-specific modifications even in complex matrices like liver or brain.
Do I need to send fresh tissue or is frozen material acceptable?
Flash-frozen tissues stored at −80°C are ideal. Please avoid multiple freeze-thaw cycles, as acylation modifications may degrade.
Which types of acylation can be quantified in a single run?
We support multiplexed detection of lysine acetylation, propionylation, succinylation, crotonylation, malonylation, and more. Co-analysis feasibility depends on the enrichment strategy selected.
What is the required amount of protein for each sample?
We recommend 5–10 mg total protein per sample. For challenging matrices or very low input amounts, please consult us to explore feasibility.
Can you perform label-based quantification like TMT or SILAC for comparative acylation profiling?
Yes. Our platform supports TMT, SILAC, and label-free workflows depending on your experimental design and throughput needs.
Will you provide site-level acylation quantification and bioinformatics interpretation?
Yes. Our deliverables include site-level quantification tables, summary statistics, pathway enrichment, and functional annotations relevant to your biological context.
Learn about other Q&A.
Client Case Study

Site-Specific Profiling of H2BK120 Acetylation Reveals HDAC4's Role in DNA Repair
Client Publication:
Di Giorgio E. et al., Nucleic Acids Research, 2024.
DOI: 10.1093/nar/gkae501
- Background
- Contribution
- Key Findings
- Conclusion & Impact
Histone acetylation is a key chromatin modification involved in the regulation of DNA repair and cell fate. While HDAC4 is known as a class IIa histone deacetylase, its specific contribution to homology-directed repair (HDR) and histone regulation remained unclear.
This study aimed to determine whether HDAC4, in coordination with HDAC1 and HDAC2, modulates H2BK120 acetylation (H2BK120ac) to regulate the recruitment of DNA repair machinery and prevent premature cellular senescence.
Creative Proteomics supported this project by performing mass spectrometry-based analysis of histone acetylation, with a focus on the H2BK120 site. Our services included:
- Sample processing and protein digestion of histones extracted from HeLa cells under various experimental conditions (wild-type, HDAC4 knockout, and rescue).
- High-resolution LC-MS/MS analysis to detect and validate acetylated peptides, including those carrying the H2BK120ac modification.
- Qualitative profiling to confirm the presence and relative change in H2BK120ac under HDAC4-deficient and rescued states.
- Mass spectrometry and immunoblotting demonstrated that H2BK120ac levels increased significantly upon HDAC4 loss.
- HDAC4 re-expression restored H2BK120ac to baseline levels.
- Functional downstream effects included reduced CtIP and BRCA1 recruitment to double-strand breaks, impairing homologous recombination-mediated repair (see Figures 3B–4A of the paper).
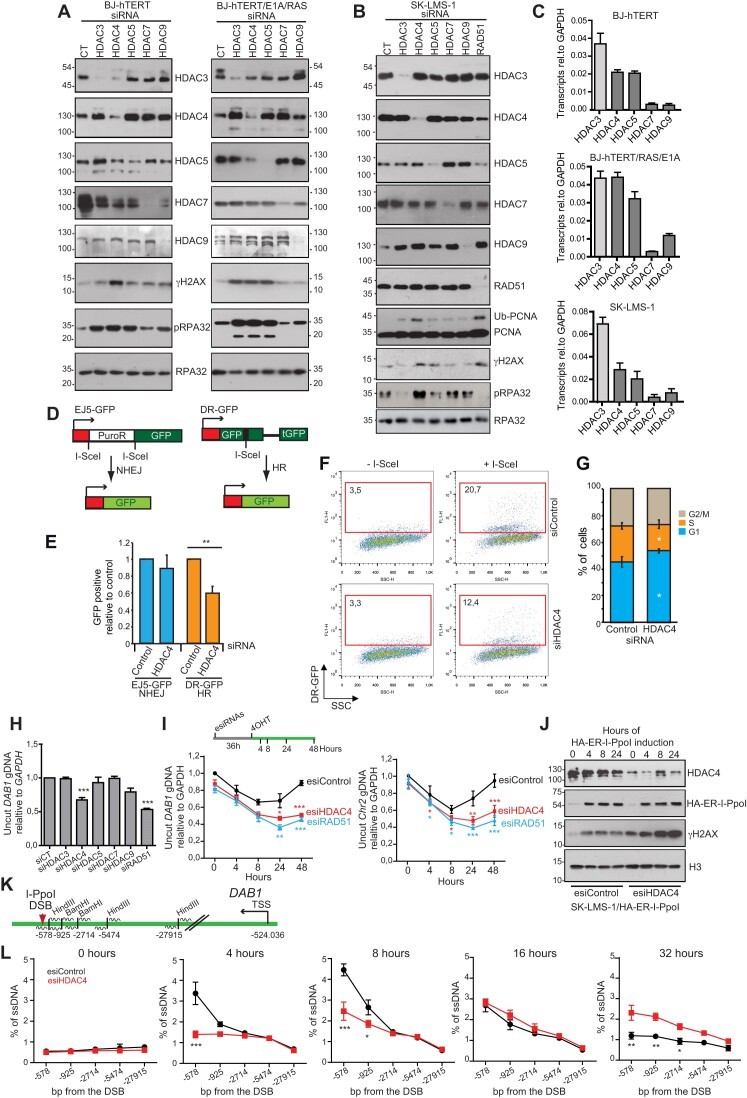 Figure. Loss of HDAC4 increases H2BK120 acetylation and disrupts recruitment of DNA repair proteins. (A–C) Western blot and immunofluorescence analysis showing elevated levels of H2BK120ac in HDAC4 knockout cells, with restoration upon HDAC4 re-expression. Quantitative analysis confirms that increased acetylation is correlated with impaired recruitment of CtIP and BRCA1 to DNA double-strand breaks.
Figure. Loss of HDAC4 increases H2BK120 acetylation and disrupts recruitment of DNA repair proteins. (A–C) Western blot and immunofluorescence analysis showing elevated levels of H2BK120ac in HDAC4 knockout cells, with restoration upon HDAC4 re-expression. Quantitative analysis confirms that increased acetylation is correlated with impaired recruitment of CtIP and BRCA1 to DNA double-strand breaks.
Creative Proteomics' site-specific histone acetylation profiling enabled the client to quantify key chromatin modifications with high confidence, providing molecular insight into HDAC4-mediated genome regulation. The study successfully linked a single lysine acetylation event (H2BK120ac) to broad changes in DNA repair capacity and senescence control.
This case highlights our platform's ability to support low-abundance, site-targeted PTM analysis in chromatin biology, epigenetics, and DNA damage response research.
Publication
Here are some publications in PTMs Proteomics research from our clients:

- Trypanosoma cruzi DNA Polymerase β Is Phosphorylated In Vivo and In Vitro by Protein Kinase C (PKC) and Casein Kinase 2 (CK2).Cells (2022). DOI: https://doi.org/10.3390/cells11223693
- Synergistic Combined-proteomics Guided Mapping strategy identifies mTOR mediated phosphorylation of LARP1 in nutrient responsiveness and dilated cardiomyopathy. bioRxiv (2022). DOI: https://doi.org/10.1101/2022.10.13.512080
- The Interplay between GSK3β and Tau Ser262 Phosphorylation during the Progression of Tau Pathology. International Journal of Molecular Sciences (2022). DOI: https://doi.org/10.3390/ijms231911610
- Exploratory phosphoproteomics profiling of Aedes aegypti Malpighian tubules during blood meal processing reveals dramatic transition in function. PLOS ONE (2022) DOI: https://doi.org/10.1371/journal.pone.0271248










