Crotonylation Analysis Services
Originally identified on histones, lysine crotonylation has now been detected in a wide range of non-histone proteins, revealing broader regulatory functions across the proteome. To uncover these roles, we offer a comprehensive crotonylation proteomics analysis service that combines:
High-specificity antibody enrichment: Using proprietary antibodies that selectively bind crotonylated peptides, we significantly enhance detection rates.
Advanced LC-MS/MS platforms: Our high-resolution liquid chromatography-tandem mass spectrometry technology enables site-level identification of crotonylated residues across thousands of proteins.
This workflow allows researchers to map crotonylation networks at an unprecedented depth—opening new doors in epigenetics, disease biomarker discovery, and drug development.
Submit Your Request Now
×
Every project with us ends in actionable insights—not just raw data. Our comprehensive crotonylation proteomics analysis package includes both experimental documentation and high-value bioinformatics interpretation.
You'll receive:
- A full laboratory report outlining the complete experimental workflow and summarising protein identification and quantification results.
- Detailed bioinformatics analysis, customised to your study goals and sample type.
- What is
- Service Details
- Publication
- Case Study
What is Crotonylation
Crotonylation—an emerging post-translational modification—is gaining traction for its pivotal role in gene regulation, cell metabolism, and chromatin dynamics. Particularly in histones, crotonylation alters how DNA interacts with regulatory proteins, making it a crucial focus in advanced proteomics research.
In our recent studies with biopharma clients, crotonylation was found to improve transcription factor accessibility. How? The addition of negatively charged crotonyl groups neutralises the positive charge of histones, loosening their grip on DNA and creating a more open chromatin structure. This promotes gene activation, especially for genes tied to reproductive cell development during mid-meiosis.
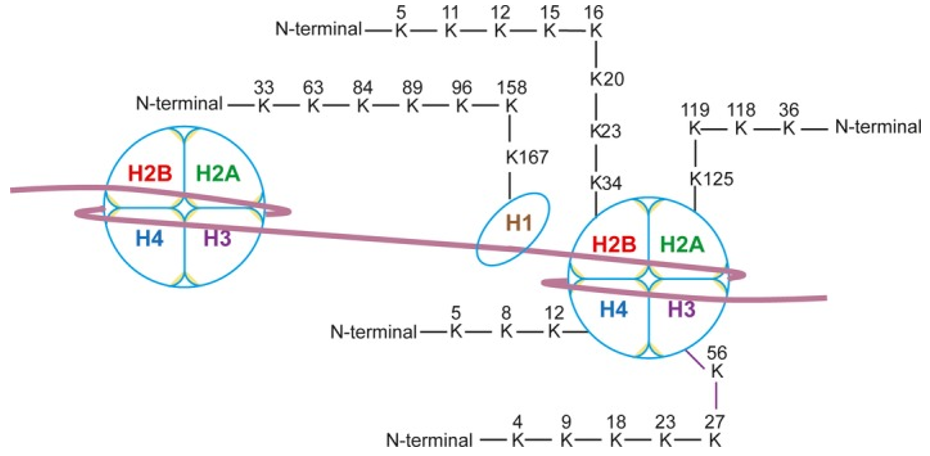 Fig. 1 Human histone crotonylation sites[1]
Fig. 1 Human histone crotonylation sites[1]
Why It Matters for Drug Developers and CROs
Whether you're refining monoclonal antibody production tips or investigating new epigenetic targets, crotonylation profiling can reveal key protein modifications driving cell behaviour. Our platform is already supporting biologics developers in uncovering novel regulatory pathways that influence therapeutic performance and cellular health metrics.
Unlock Functional Protein Insights with Our Crotonylation Proteomics Service
At Creative Proteomics, we offer a specialised crotonylation proteomics service tailored for complex biological samples—including tissues and cell lines. Designed for drug developers and academic labs alike, this service leverages cutting-edge mass spectrometry to pinpoint both crotonylated proteins and their precise modification sites.
Workflow of service
Customized Bioinformatics Services
| Standard Analysis | Customized Analysis |
|---|---|
| a) Collection of output statistics b) Establishment of database and search for modified proteins c) Identification of modified proteins and modification sites d) Annotation of modified proteins e) Quantitative analysis of modified proteins f) Analysis of differential expression of modified proteins g) Functional enrichment analysis of differentially modified proteins GO/KEGG | Modified peptide motif analysis Protein network interaction prediction Protein domain prediction (Advanced Analysis) Protein subcellular localization analysis (Advanced Analysis) |
What Sets Our Platform Apart
High-resolution mass spectrometry: Delivers accurate, site-specific data on crotonylated peptides.
Expert bioinformatics support: Our team helps interpret the functional relevance of your crotonylation profiles.
End-to-end project handling: From sample prep to final report, we manage every step with scientific rigour.
High-Specificity Enrichment Meets Quantitative Mass Spectrometry
Our crotonylation proteomics platform combines chemical stability with technical precision—offering a reliable solution for researchers exploring protein regulation, disease mechanisms, and therapeutic targets.
Targeted peptide enrichment: We use chemically stable crotonyl-specific resins for immunoprecipitation (IP), ensuring high specificity and strong enrichment efficiency.
Advanced mass spectrometry: Enriched crotonylated peptides are identified at scale using high-resolution, high-scan-speed MS technology.
Quantitative comparison: Compatible with common quantitation methods (e.g., TMT, SILAC), allowing researchers to measure differences in crotonylation levels across multiple samples.

Thermo Q ExactiveTM series

AB Sciex 6500+

Thermo Orbitrap Fusion Lumos

Bruker timsTOF Pro
Ideal for Drug Discovery and Functional Protein Research
Our platform supports a wide range of applications in both academic and industry settings:
Target identification for drug development
Screening novel disease biomarkers
Decoding molecular mechanisms of action
Constructing proteomic blueprints for specific species
To ensure accurate mapping and meaningful insights, we recommend that your project be based on a species with a well-annotated genome, EST data, or complete protein sequence library.
Sample Requirements for Crotonylation Analysis
| Sample Type | Minimum Amount Required | Additional Notes |
|---|---|---|
| Fresh animal tissue | ≥ 600 mg | |
| Fresh plant tissue | ≥ 6 g | |
| Cell culture | ≥ 1 × 10⁷ cells/tube × 3 tubes | |
| Fungi and bacteria | ≥ 600 mg | |
| Serum or plasma | 450 μL × 4 tubes | |
| Protein solution | Total protein of 5–10 mg | |
| Urine (body fluid) | 15 mL × 4 tubes | Centrifuge at 1000 × g for 5 minutes; discard sediment |
| Other body fluids | > 15 mL | Includes saliva, amniotic fluid, cell culture supernatant, etc. |
Publication
Here are some publications in PTMs Proteomics research from our clients:

- In Vitro Identification of Phosphorylation Sites on TcPolβ by Protein Kinases TcCK1, TcCK2, TcAUK1, and TcPKC1.Microorganisms (2024) DOI: https://doi.org/10.3390/microorganisms12050907
- Tumor-Derived Extracellular Vesicles Regulate the Fate and Function of T-Lymphocyte Via Reprogramming Lipid Metabolism. Institution: Saint Louis University (Dissertation, 2023). https://search.proquest.com/openview/8ee9ca786d08344a371a238fa15f7375
- Exploratory phosphoproteomics profiling of Aedes aegypti Malpighian tubules during blood meal processing. Journal: PLOS ONE (2022). DOI: https://doi.org/10.1371/journal.pone.0271248
- The Interplay between GSK3β and Tau Ser262 Phosphorylation during the Progression of Tau Pathology. Journal: International Journal of Molecular Sciences (2022) DOI: https://doi.org/10.3390/ijms231911610
Case Study

Proteomics analysis of lysine crotonylation and 2-hydroxyisobutyrylation reveals significant features of systemic lupus erythematosus. Clin Rheumatol. 2022 Dec;41(12):3851-3858. doi: 10.1007/s10067-022-06254-4
- Objective
- Methods
- Results
- Conclusion
This study aimed to investigate the patterns and biological significance of two lysine post translational modifications—crotonylation (Kcr) and 2 hydroxyisobutyrylation (Khib)—in peripheral blood mononuclear cells (PBMCs) from patients with systemic lupus erythematosus (SLE), with the goal of identifying key modified proteins and pathways that may underpin SLE pathogenesis and could serve as potential biomarkers.
- Sample preparation: PBMCs were collected from SLE patients and healthy controls.
- Protein extraction & peptide labelling: Proteins were extracted, digested, and labeled using tandem mass tags (TMT). Modified peptides carrying Kcr and Khib were enriched using specific antibodies.
- LCMS/MS analysis: Enriched peptides were subjected to high-resolution liquid chromatography–tandem mass spectrometry (LCMS/MS) for quantitative profiling.
- Bioinformatics:
- Identified proteins dually modified by both Kcr and Khib ("dual modified proteins", DMPs);
- Constructed protein–protein interaction (PPI) networks;
- Performed Gene Ontology (GO) and KEGG pathway enrichment analyses to elucidate functional implications.
- A total of 76 DMPs (proteins with both Kcr and Khib modifications) were significantly altered in SLE versus healthy controls.
- GO enrichment indicated strong involvement in MHC class II protein complex binding (antigen presentation) and leukocyte migration.
- KEGG analysis highlighted two central pathways:
- Antigen processing and presentation: Six DMPs (CLTC, HSPA1B, HSPA8, HSP90AB1, HSPD1, PDIA3), with HSPA8 as the hub. The increased Kcr/Khib on HSPA8 suggests enhanced ATP hydrolysis and antigen-loading to MHC II.
- Leukocyte transendothelial migration: Seven DMPs including ACTN1, ACTN4, EZR, MSN, RAC1, RHOA, and VCL. Notably, MSN exhibited the highest number of modification sites; alterations in its N-terminal FERM domain may strengthen leukocyte–endothelial adhesion, a key migration step.
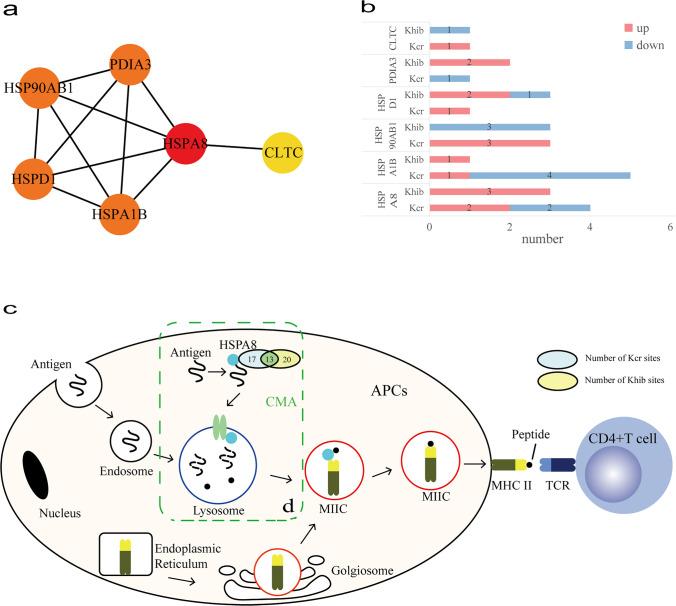 Figure 2: Displays the PPI network of DMPs in the antigen processing and presentation pathway, highlighting HSPA8 and its modification sites.
Figure 2: Displays the PPI network of DMPs in the antigen processing and presentation pathway, highlighting HSPA8 and its modification sites.
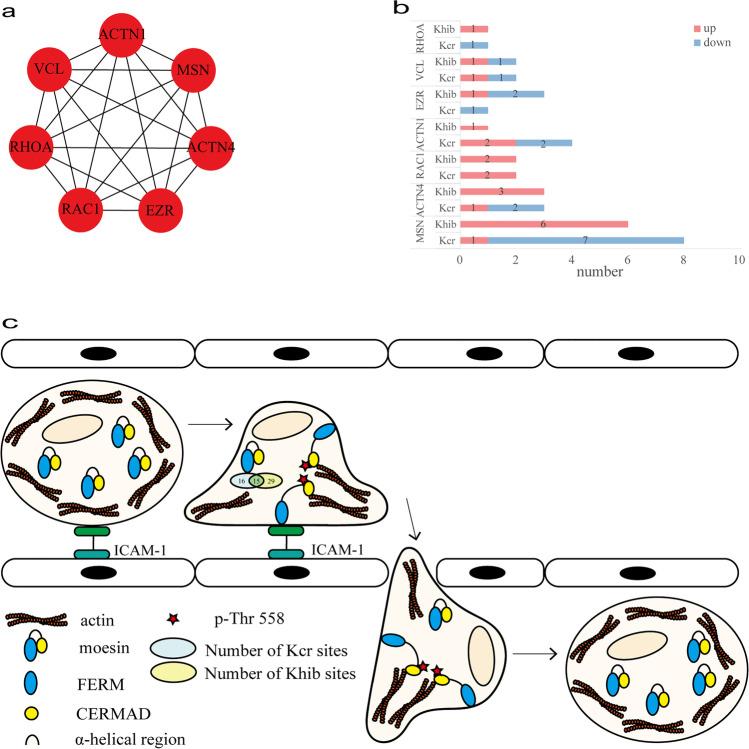 Figure 3: Illustrates the PPI network of DMPs in leukocyte transendothelial migration, emphasizing proteins such as MSM, ACTN1/4, and RAC1 with modification site counts.
Figure 3: Illustrates the PPI network of DMPs in leukocyte transendothelial migration, emphasizing proteins such as MSM, ACTN1/4, and RAC1 with modification site counts.
The coordinated upregulation of Kcr and Khib modifications appears to enhance both antigen presentation and leukocyte extravasation, indicating a synergistic mechanism that may contribute to immune activation and tissue damage in SLE. These PTMs therefore represent promising targets for further investigation into SLE pathogenesis .
References
- Junhu Wan, Hongyang Liu, Jie Chu, Hongquan Zhang. Functions and mechanisms of lysine crotonylation. J Cell Mol Med. 2019 Nov; 23(11): 7163–7169. DOI: 10.1111/jcmm.14650
- Gaoyue Jiang, Chunxia Li, Meng Lu, Kefeng Lu & Huihui Li Protein lysine crotonylation: past, present, perspective. Cell Death & Disease 12, 703 (2021). DOI: 10.1038/s41419-021-03987-z
- Shuo Wang1, Guanqun Mu1, Bingquan Qiu1, Meng Wang1, Zunbo Yu2, Weibin Wang3, Jiadong Wang3, Yang Yang. The Function and related Diseases of Protein Crotonylation. Int J Biol Sci 2021; 17(13):3441-3455. PMID: 34512158














