The glycan chain is located in the outermost layer of the cell and is the first molecule encountered and recognized by other cells, antibodies, invading viruses and bacteria. N-glycans regulate a wide range of cellular functions, including cell-cell and cell-matrix adhesion, cell proliferation, cell survival and immune system response, by affecting protein folding, recognition, clearance, secretion and construction.
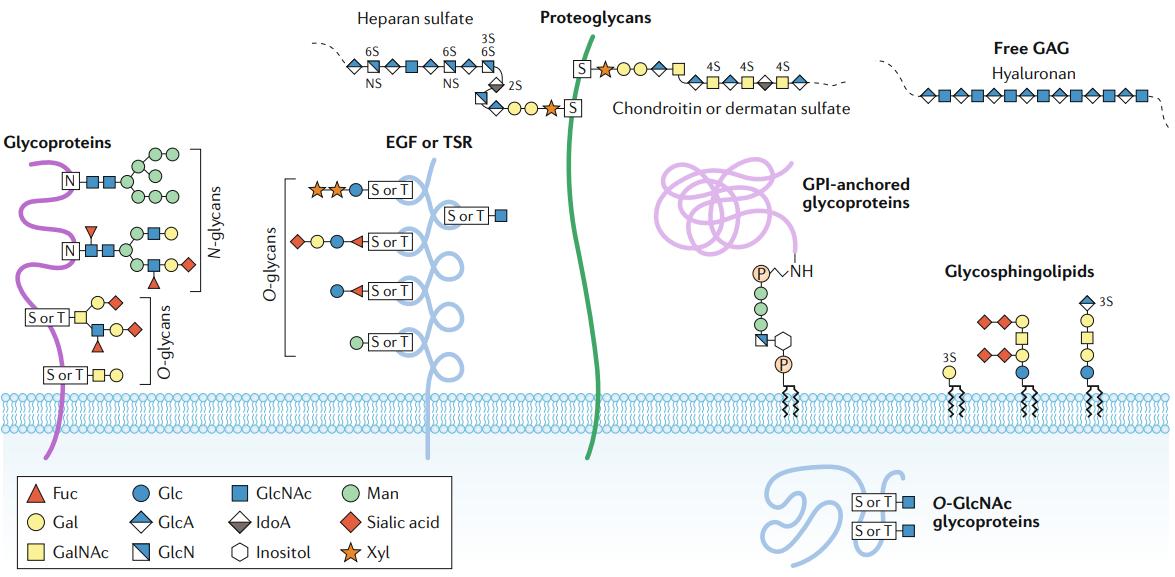
Major types of glycosylation in humans (Reily et al., 2019)
The difficulty in N-glycosylation analysis lies in identifying the glycosylation modification sites of proteins and in analyzing the highly complex N-glycans attached to the glycosylation sites. N-glycosylation modifications are not guided by a template and are performed by multiple enzymes acting together. It is highly influenced by different physiological conditions, and multiple glycosylation sites may exist on a single protein. Moreover, the same glycan chain modification can occur at multiple glycosylation sites, which is called macroheterogeneity of protein glycosylation. In addition, different glycan modifications can occur at the same glycosylation site, which is called microheterogeneity of glycosylation. Complex glycan chains can form stereo-isomers and regio-isomers. A comprehensive study of these targets can make the analysis exceptionally complex.
N-glycosylation Analysis Strategies
(1) Intact analysis: MS analysis of intact glycoproteins under denaturing or non-denaturing conditions. This method allows to obtain complete protein information of glycoproteins as well as to directly analyze the combination of post-translational modifications and modifications. This technique is challenging due to the large microhomogeneities exhibited by glycoprotein structures and the limitations of existing techniques to effectively separate glycoproteins from proteins.
(2) Bottom-up glycoproteomic analysis: The glycan chains and peptide chains of glycoproteins are analyzed separately. The released N-glycans are isolated and structurally analyzed. In addition, the digestion of glycoproteins into glycopeptides allows the study of specific glycosylated content using a peptide-centered proteomic analysis. The advantage of this method is that the specific site of glycosylation on the protein can be identified and that site-specific information can be used to identify different glycosylation sites for a specific glycan structure. It also helps to understand the molecular structure of the protein.
N-glycan Analysis Procedures
- N-glycan release
The main methods for N-glycan release are enzymatic and chemical.
An earlier and more effective method for the chemical release of N-glycans is hydrazinolysis. However, anhydrous hydrazine is a highly toxic and explosive solvent and the released N-glycans under such conditions are subject to further degradation and isomerization, resulting in inhomogeneity of the sugar chains and causing some interference in the subsequent analysis. Therefore, the use of hydrazinolytic methods has been greatly reduced and replaced by milder enzymatic methods.
Glycosidases are a powerful tool in N-glycan analysis and are divided into exonucleolytic and endonucleolytic glycosidases. The release of N-glycans from glycoproteins using the endo-glycosidase method is the most direct and the simplest. Endoglycosidases can cut intact N-glycans from peptide chains or hydrolyze the glycosidic bonds within N-glycans, cutting long glycans into shorter oligosaccharide fragments for easy analysis.
Among the various endoglycosidases, peptide-N-glycosidase F (PNGase F) is commonly used. PNGase F is generally effective in releasing the N-glycan structures of glycoproteins in the mammalian system, with the exception of structures with core α1-3 rockulose attached to the reducing terminus GlcNAc. In this case PNGase A is effective for all structures. Glycoside endonucleases (Endo) are another class of endoglycosidases used for N-glycan release. Unlike PNGase F, this enzyme specifically releases the glycosidic bond between the two GlcNAc structures in the core pentasaccharide structure of the N-glycan. Any structure attached to the reducing terminal GlcNAc residue is retained (e.g., rockulose, peptides linked by glycosylation sites). Due to the specificity of Endo function, it can be used complementarily to PNGase F binding in N-glycan release studies.
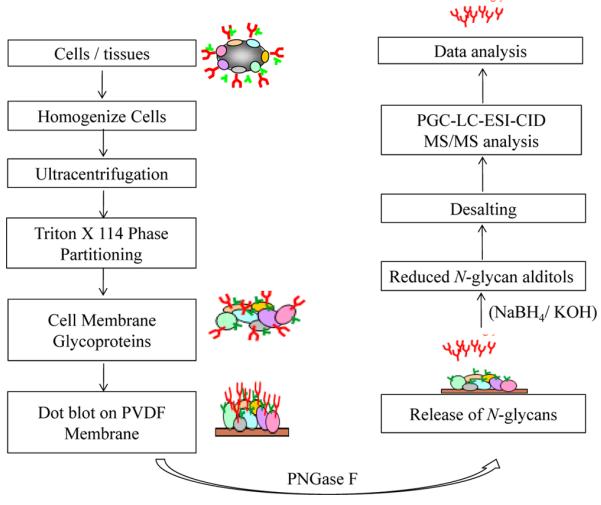
Schematic diagram of the protocol for enrichment of membrane glycoproteins, followed by release and processing of N glycans for LC-MS/MS analysis (Sethi et al., 2016).
- N-glycan labeling
Free N-glycans lack chromophore or fluorophore properties, making chromatography-based analysis difficult. N-glycans are also not ideal analytes for MS. Its strong hydrophilicity leads to inefficient desolvation, resulting in suboptimal ionization efficiency during MS analysis. Therefore, chemical labeling (i.e. derivatization) of N-glycans is often used prior to analysis to modulate the physicochemical properties of N-glycans and increase the sensitivity of the assay.
N-glycan derivatization can be divided into four main types: (1) holomethylation modification, (2) reducing end modification, (3) sialic acid modification, and (4) multiple labeling modification of N-glycans.
Holomethylation is one of the most commonly used methods in N-glycan derivatization reactions. It forms a methanol salt with the hydroxyl group on the N-glycan, a carboxyl ester carboxylate with the sialic acid residue, and adds a methyl group to the aldehyde group at the reducing end. After holomethylation, the detection sensitivity of the derivatives is significantly improved compared to the non-derivatized N-glycans. In addition, the conversion of the acidic structure into a neutral structure allows the simultaneous detection and analysis of different types of N-glycan derivatives in positive ion mode.
Reduced-end modifications of N-glycans have been investigated in various aspects, including reductive amination, glycosamine labeling, hydrazide labeling, Michael addition, etc., to adapt N-glycan derivatives to various analytical techniques. The two main types are the addition of fluorescent labels and charged group labels, which are mainly used for high performance liquid chromatography (HPLC) as well as mass spectrometry (MS) analysis.
In MALDI-MS, the sialic acid in N-glycans is unstable and easily lost or partially lost, mainly due to the unstable acidic protons. Therefore, modification of the sialic acid in N-glycans prior to analysis is required to overcome this drawback. This is mainly done by forming salts or introducing derivatives that stabilize the acid.
N-glycans can be divided into two groups: neutral and acidic sugars, with large differences in ionization efficiency between them. Labeling only the reducing end of N-glycans affects the ionization efficiency of sialic acid glycans on MS. Multiple labeling of N-glycans is used to solve this problem. Commonly used sugar multiple labeling is the combination of the released N-glycan reducing end labeling (reducing amination, hydrazine derivatization, glycosamine-derivatization, etc.) with sialic acid modification (alkyl amidation, etc.) for MS analysis. Quantitative analysis can also be performed on this basis using isotope reagents.
- N-glycan instrument detection
The separation and structural analysis of N-glycans are generally performed by liquid chromatography (LC), mass spectrometry (MS), capillary electrophoresis (CE), etc.
1) Liquid chromatography analysis
Several LC separation modes have been used for the analysis of N-glycans, including hydrophilic interaction chromatography (HILIC), porous graphitized carbon chromatography (PGC), high pH anion exchange chromatography (HPAEC), and reverse liquid chromatography (RPLC). N-glycan derivatization plays an important role in these analyses in enhancing retention and detection sensitivity on the stationary phase. Liquid chromatographic methods often applied to the analysis of N-glycan derivatives are RPLC and HILIC.
2) Capillary electrophoresis analysis
Capillary electrophoresis (CE) is also commonly used for N-glycan analysis. However, some N-glycans containing acidic monosaccharides (structures such as sialic acid) and most N-glycans are uncharged and cannot be separated effectively in CE. N-glycans lack chromophores for effective optical detection. Therefore, in the CE analysis of N-glycans, chemical labeling is often used to ensure the migration and detection of N-glycan derivatives in the electric field. One of the most widely used labeling reagents for CE analysis of N-glycans is the fluorescent reagent 8-amino-1-1,3,6-trisulfonate (APTS). It can react with the reducing end of the released N-glycan with an efficiency close to 100%.
3) Mass spectrometry analysis
The mass spectra of N-glycans contain a wealth of information about the compounds. In most cases, the molecular weight, molecular formula and molecular structure can be determined by the resolution of mass spectrometry results. The amount of sample used for mass spectrometry analysis is extremely small, making mass spectrometry a powerful tool for highly sensitive analysis of N-glycans. Although there are many ionization methods used for the ionization of intact biomolecules, the two modalities used almost exclusively for N-glycan analysis are matrix-assisted laser desorption ionization (MALDI) and electrospray ionization (ESI).
MALDI-MS is well suited for the analysis of N-glycan mixtures that do not contain anionic groups. Anionic mixtures (e.g., sialylated glycans) tend to fragment during MALDI-MS, but can be stabilized by chemical methods such as sialic acid modification. MALDI-MS allows rapid and highly sensitive and high throughput analysis of N-glycans from very small amounts of samples of biological origin (e.g. cell lysates or body fluids) to facilitate clinical studies.
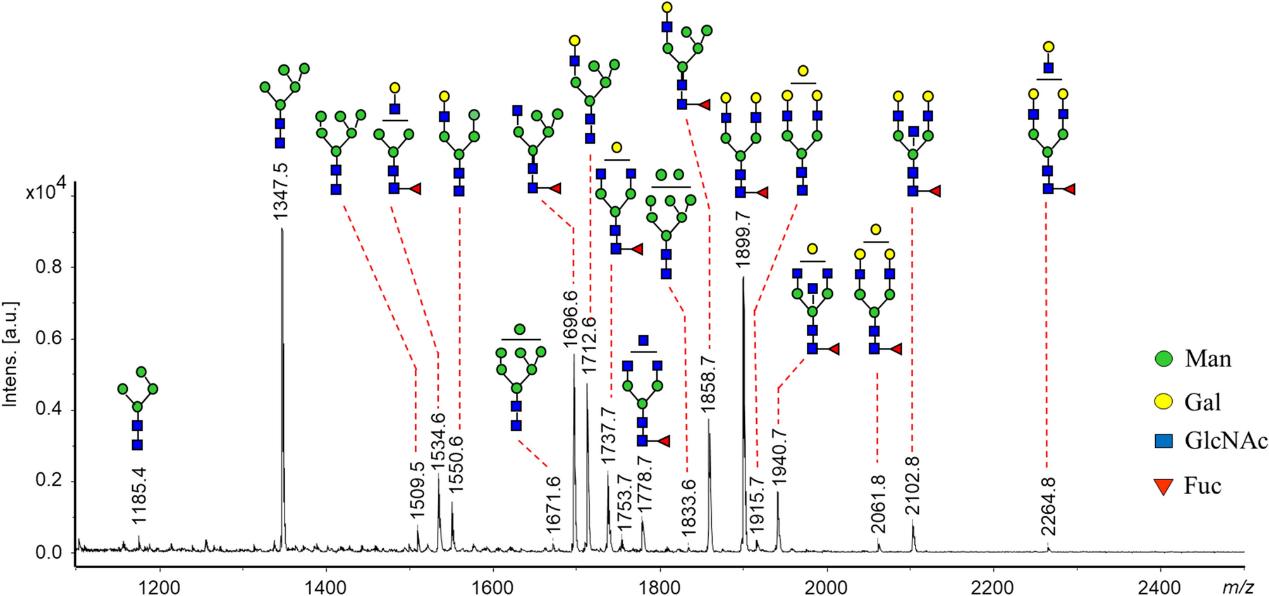
MALDI-MS spectrum of N-glycans recorded from SDS-purified E glycoprotein of TBEV grown in human neuroblastoma cells (UKF-NB4) (Lattová et al., 2020)
Different from MALDI, the analysis of N-glycan compounds using ESI produces several different types of ions, depending on the conditions of use of the ion source and the solvent additions.The ESI technique is widely used for N-glycan analysis because it can be easily used in conjunction with other techniques (e.g. LC, CE, etc.), overcomes many of the limitations of MALDI analysis, and identifies a significantly larger number of glycans.
4) Coupling techniques
LC-MS is currently the most reliable and stable method used for isomer separation and MS detection.LC-MS has several methods to analyze N-glycans depending on the separation mode of LC, mainly C18, HILIC, and PGC. C18 is still the most commonly used stationary phase. The hydrophilic nature of N-glycans results in poor retention on C18 columns, so N-glycans generally require derivatization.
- Analysis of sample data of N-glycans
For large numbers of samples, a large amount of N-glycan data needs to be statistically analyzed. If the data are normally distributed, independent sample t-test (T-test) or analysis of variance (ANOVA) can be used to determine the differences between samples and to test the data for homogeneity of variance. If the sample data have homogeneity of variance, the ANOVA results are retained and further parametric tests are performed. If the sample does not have homogeneity of variances, further analysis by non-parametric tests is required.
Discriminant analysis (DA) is used to assess the adequacy of the classification system. Unlike ANOVA or multiple ANOVA analysis, which predicts one or more continuous dependent variables by one or more independent variables, DA is a powerful analytical method to determine the validity of the prediction of a set of variables across various samples. The main data processing methods commonly used in DA are principal component analysis (PCA) or partial least squares (PLS).
Even if there is a statistical difference in one or some N-glycan data between two groups of samples with different conditions, this does not necessarily mean that this or these N-glycans can be used as a diagnostic marker to distinguish between the two. Further subject characteristic curve (ROC) analysis is required. In ROC analysis, curves are plotted based on the true positive rate (sensitivity) and false positive rate (specificity). These area under the curve (AUC) plots are then used to distinguish whether or not they can be used as biomarkers. In addition, in clinical diagnostic analysis, samples are sometimes studied in multivariate analysis in combination with multiple markers.
Reference
- Reily, C., Stewart, T. J., Renfrow, M. B., & Novak, J. (2019). Glycosylation in health and disease. Nature Reviews Nephrology, 15(6), 346-366.
- Sethi, M. K., Hancock, W. S., & Fanayan, S. (2016). Identifying N-glycan biomarkers in colorectal cancer by mass spectrometry. Accounts of chemical research, 49(10), 2099-2106.
- Lattová, E., Straková, P., Pokorná-Formanová, P., Grubhoffer, L., Bell-Sakyi, L., Zdráhal, Z., ... & Ruzek, D. (2020). Comprehensive N-glycosylation mapping of envelope glycoprotein from tick-borne encephalitis virus grown in human and tick cells. Scientific reports, 10(1), 1-10.







