Accurate Prenylation Site Mapping via LC-MS/MS
At Creative Proteomics, we specialize in comprehensive post-translational modification (PTM) analysis—offering an end-to-end prenylation site identification service designed for large-scale proteomic studies.
With our expertise in mass spectrometry-based lipid modification analysis, we streamline the entire workflow so you can focus on your research. Simply submit your biological samples—our team will handle the rest, including:
- Protein extraction and enzymatic digestion
- Peptide separation and enrichment of prenylated peptides
- High-resolution LC-MS/MS analysis
- Raw data processing and in-depth bioinformatics interpretation
Whether you're studying signaling pathways, cancer-related targets, or microbial mechanisms, our prenylated proteome service delivers high-confidence site identification with actionable insights.
Submit Your Request Now
×
- Experimental procedures
- Mass Spectrometry Parameters
- Details of prenylation
- Mass Spectrogram
- What is
- Service Details
- Delivery & Demo
- FAQ
What is Protein Prenylation
Prenylation is a specialized post-translational lipid modification (PTMs) that helps proteins attach to cell membranes. This process plays a critical role in regulating signal transduction, especially in diseases such as cancer and microbial infections.
Why Prenylation Matters
Proteins modified by prenylation—typically through farnesyl or geranylgeranyl isoprenoids—are often directed to cellular membranes. This membrane localization is essential for their biological function, especially in signaling pathways. For instance:
- Prenylated proteins contribute to cell communication and signal relay.
- Many participate in pathways governing growth, survival, or immune response.
- The isoprenoid tail helps them anchor into the lipid bilayer, a requirement for proper protein positioning and function.
While most known effects of prenylation relate to membrane attachment, researchers are increasingly exploring its influence on protein-protein interactions. In some cases, experimental evidence suggests the lipid modification itself enhances or stabilizes key molecular contacts—beyond just helping the protein find the membrane.
Therapeutic Potential: Targeting Prenylation in Drug Discovery
One of the earliest breakthroughs in this field came from studying Ras proteins, a well-known family involved in regulating cell proliferation. Mutant Ras proteins—implicated in many cancers—require farnesylation to function abnormally and drive tumor growth.
This finding sparked intense interest across pharmaceutical R&D in prenylation inhibitors. By blocking this lipid tagging process, it's possible to interfere with the localization and activity of disease-related proteins—offering a novel therapeutic strategy.
In fact, several research groups now focus on developing small molecules that selectively inhibit prenyltransferases, the enzymes responsible for prenylation.
Mapping the Prenylated Proteome with Chemical Tools
To understand how prenylation impacts cellular function, scientists need to identify which proteins are prenylated—and when. Traditional methods often lacked sensitivity or specificity. That's where chemical proteomics steps in.
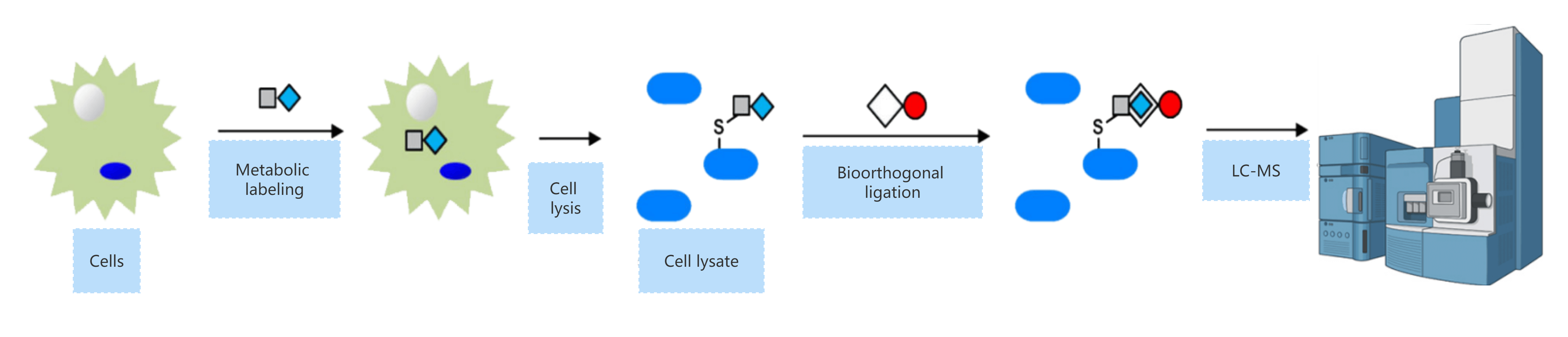
Here's how modern labeling-based techniques work:
Metabolic tagging: Living cells are incubated with isoprenoid analogs that contain a chemical "handle" such as an azide or alkyne group.
Bioorthogonal labeling: These tags are later clicked onto reporter molecules like fluorophores (for imaging) or biotin (for enrichment and mass spectrometry).
Protein analysis: The labeled proteins are visualized on gels or pulled down and identified using LC-MS/MS, giving researchers a detailed view of the prenylated proteome under physiological conditions.
These advanced workflows allow for high-resolution, system-wide mapping of prenylated proteins—shedding light on their dynamics in cancer, infection, and other cellular contexts.
Our Prenylation Site Identification Service
At Creative Proteomics we combine advanced mass spectrometry with customized analysis to offer a leading prenylation analysis service. Our expertise lies in accurately identifying and quantifying key protein modifications—farnesylation and geranylgeranylation.
This precise profiling empowers researchers and drug developers to better understand how these lipid modifications regulate cellular functions and contribute to disease mechanisms. With our reliable service, you gain insightful data that supports your research on protein prenylation's biological roles and therapeutic potential.
Integration with Other PTM Analyses
Our platform enables simultaneous or sequential enrichment of multiple PTMs. We offer a comprehensive, one-stop service covering sample preparation, in-depth peptide separation, and mass spectrometry detection. This allows for the quantification of various PTMs within the same experiment.
By profiling several modifications together, we can build detailed PTM interaction networks. These networks reveal whether different modifications coexist on a protein or occur exclusively. For example, detecting both phosphorylation and prenylation sites on the same protein sheds light on how these modifications may cooperate or oppose each other. Such insights deepen our understanding of cellular regulation and signalling pathways.
This integrative approach helps researchers uncover complex layers of protein regulation that single-modification analyses might miss. It supports more comprehensive studies of protein function and disease mechanisms.
Advantages of Our Prenylation Site Identification Service
- Broad Application Scope: Perform both qualitative and relative quantitative analyses of diverse protein prenylation sites with consistent and trustworthy results.
- User-Friendly Workflow: Avoid complex steps like radioisotope labeling or Edman degradation sequencing, making the process straightforward and efficient.
- Enhanced Site Coverage (Optional): Utilize labeling of prenylated peptides to boost detection sensitivity and maximize coverage of modification sites.
- High Sensitivity Analysis: We use state-of-the-art mass spectrometry to detect even low-abundance prenylated proteins with great accuracy.
- Efficient and High-Throughput: Our platform handles large sample volumes efficiently, making it ideal for high-throughput projects.
- Accurate and Reliable Data: We deliver comprehensive, reproducible results to support confident interpretation and further study.
Workflow of Prenylation Site Identification
Target Protein Labeling
Depending on your sample type, we label the prenylated proteins or peptides to enhance detection sensitivity.
Isoprenylated Peptide Enrichment (Optional)
For samples with low abundance of prenylated peptides, we apply selective enrichment to improve identification rates.
HPLC Separation and MS/MS Analysis
Peptides are separated using high-performance liquid chromatography (HPLC) and then analyzed by tandem mass spectrometry (MS/MS) for detailed characterization.
Prenylation Site Data Analysis
Advanced bioinformatics tools interpret the mass spectrometry data to accurately map prenylation sites.
Bioinformatics Analysis Support
Beyond peptide identification and site mapping, we offer extensive bioinformatics support, including:
- Gene Ontology (GO) annotation
- KEGG pathway enrichment
- COG/KOG functional classification
- Protein domain annotation
- Subcellular localization prediction
These analyses reveal potential roles of prenylated proteins in cellular processes and signaling pathways. We also build protein-protein interaction and modification networks to explore regulatory pathways. Results are presented as tables and figures highlighting enriched biological processes and interaction modules, facilitating comprehensive biological interpretation.
Service Applications of Prenylation Site Identification
Cancer Research: Investigate the prenylation mechanisms of Ras and other oncogenic proteins to better understand tumor progression.
Neurodegenerative Diseases: Analyze prenylation abnormalities to reveal insights into disease development and pathology.
Drug Development: Support the design and evaluation of therapeutics targeting prenylation pathways.
Protein Function Studies: Examine protein modifications and their functional roles across various species and biological systems.
Sample Requirements
| Sample Type | Required Amount | Additional Notes |
|---|---|---|
| Fresh Animal Tissue | ≥ 600 mg | |
| Fresh Plant Tissue | ≥ 6 g | |
| Cell Culture | ≥ 1 × 10⁷ cells per tube × 3 tubes | |
| Fungi and Bacteria | ≥ 600 mg | |
| Serum, Plasma | 450 μL × 4 tubes | |
| Protein Solution | Total protein 5–10 mg | |
| Body Fluid Samples | Urine: 15 mL × 4 tubes | Centrifuge at 1000 × g for 5 min; discard sediment |
| Other fluids (saliva, amniotic fluid, cell culture supernatant, etc.) | ≥ 15 mL |
These are general references; we can customize sample amounts according to your project needs. To preserve sample integrity, freeze samples rapidly after collection (avoid repeated freeze-thaw cycles) and ship on dry ice to minimize protein degradation and ensure high-quality data.
Data Deliverables
Our service delivers comprehensive identification and quantification of prenylated proteins in your samples, along with detailed visualization and a written report. Typical outputs include:
- Lists of identified prenylated peptides and modification sites, with corresponding protein IDs
- Quantitative or relative abundance data
- Experimental methods and raw data (LC-MS/MS results)
- Bioinformatics analyses such as pathway enrichment and functional annotation
- Visual charts (heatmaps, enrichment bar plots, interaction networks)
- A summary report covering methods, results, and conclusions
DEMO Illustration

Figure: Representative DEMO Illustration of Protein Prenylation Profiling Results
(For demonstration purposes only; not actual experimental data)
Frequently Asked Questions
What research areas benefit most from prenylation profiling? What scientific value does it provide?
Protein prenylation is a lipid modification essential for many small GTPases like Ras family proteins, facilitating their correct membrane localization. This modification regulates key signaling pathways critical for cell growth, differentiation, and survival. Aberrant prenylation is linked to diseases such as cancer, inflammation, and aging. For instance, Ras-driven cancers depend on prenylation for tumor growth, making it a target for anti-cancer drugs that inhibit prenylation enzymes.
Key applications of prenylation profiling include:
- Target discovery and validation: Identify proteins with altered prenylation in disease models or after drug treatment to find novel therapeutic targets
- Mechanistic studies: Clarify how prenylation affects protein activity, localization, and signaling pathways
- Drug development: Assess effects of inhibitors (e.g., farnesyltransferase inhibitors) on prenylated proteins, supporting drug mechanism and target validation
- Biomarker identification: Discover prenylation-modified proteins as potential disease biomarkers
Overall, prenylation proteomics offers precise protein modification data valuable for biomedical research, particularly in cancer, neurodegeneration, and cardiovascular diseases.
How do you ensure sample data privacy and intellectual property protection?
We strictly protect client data and intellectual property. All samples and data are used solely for the client's specified research purposes and are never disclosed or shared without permission. Confidentiality agreements (NDAs) are available upon request. Analysis results and reports are provided exclusively to the client. You may request the return or destruction of leftover samples after completion. Our protocols comply with industry standards and ethical guidelines, ensuring a secure and trustworthy service environment.
What advantages does your prenylation profiling service have over competitors?
Our service offers several distinct advantages:
- Comprehensive site coverage: Combining multiple enrichment and labeling techniques enables detection of a wider variety of prenylation sites, including low-abundance modifications. For example, Creative Proteomics emphasizes enhanced detection sensitivity for extensive site coverage. Creative Proteomics has quantitatively analyzed over 80 different modified proteins.
- High sensitivity and throughput: Utilizing state-of-the-art mass spectrometry and chemical labeling allows precise identification of low-level prenylated proteins, supporting large-scale projects.
- Broad applicability: Our platform works across diverse sample types and species, including mammalian cells, animal models, plants, yeast, and bacteria.
- Mature and stable platform: Experienced teams and rigorous quality control ensure reproducible, reliable data from sample prep through analysis.
In summary, our cutting-edge techniques and expert team deliver higher sensitivity, richer modification information, and deeper bioinformatics insights to accelerate your research breakthroughs.
Learn about other Q&A about other technologies.
Case Analysis: In Vivo Brain Prenylomic Profiling in an Alzheimer's Transgenic Mouse Model

- Background & Objective
- Methodology & Workflow
- Key Findings
- Biological & Disease Relevance
- Technical Strengths & Innovation
- Conclusion
Protein prenylation—lipid attachment via farnesyl or geranylgeranyl transfer—is known to regulate protein localization, trafficking, and signal transduction. Dysregulation in Alzheimer's disease (AD) has been suspected, but direct in vivo quantification was lacking. This study aimed to systematically profile prenylated proteins ("prenylome") in the brains of APP/PS1 transgenic AD mice versus wild-type (WT) controls using metabolic chemical probing coupled with proteomics.
2.1 In Vivo Metabolic Labeling
Probe used: C15AlkOPP, an alkyne-modified isoprenoid analogue compatible with both farnesyltransferase and geranylgeranyltransferase.
Delivery: 13-day intracerebroventricular (ICV) infusion to ensure widespread brain labeling.
2.2 Click-Enrichment & Proteomic Analysis
Post infusion, brains were harvested and subjected to click-chemistry tagging (e.g., biotin or fluorophore), affinity enrichment, and tryptic digestion.
Labeled peptides were analyzed using LC MS/MS and quantified with TMT labeling and MaxQuant processing. Non-prenylated peptides were filtered out .
Total prenylated proteins identified: 36 enriched in APP/PS1 brains.
Significantly upregulated prenylated proteins: 15 consistently elevated across biological replicates.
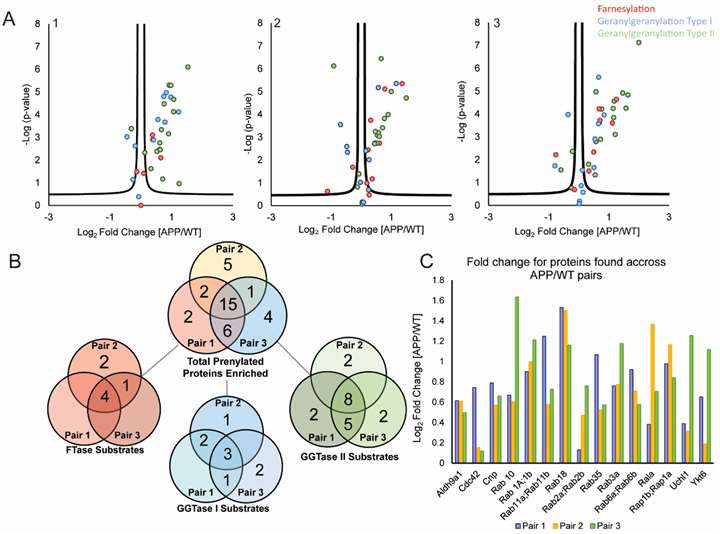 Differential prenylation in APP/PS1 vs. WT mouse brains. Three independent volcano plots (FDR = 5%) show proteins enriched in transgenic samples, and the accompanying bar graph quantifies the fold-change for the 15 common hits.
Differential prenylation in APP/PS1 vs. WT mouse brains. Three independent volcano plots (FDR = 5%) show proteins enriched in transgenic samples, and the accompanying bar graph quantifies the fold-change for the 15 common hits.
Among the 15 proteins, several are AD-related, including Rab GTPases and other trafficking proteins. Pathway analysis revealed connections to amyloid precursor protein (APP) and tau networks.
The observation that prenylation changes exceed changes in total protein abundance or transcript levels (supported by RNA seq and proteomic comparisons) implies post-translational regulation is a significant feature in AD pathology .
- In vivo metabolic labeling allows physiologically relevant measurement, avoiding artifacts from cell culture systems.
- Using LC MS/MS in combination with click chemistry enables specific, quantitative detection of prenylated peptides.
- The robust engineering of C15AlkOPP delivery ensures effective in-brain labeling across extended time periods.
This study exemplifies a state-of-the-art proteomics pipeline detecting disease-associated post-translational modifications in an in vivo AD model. The findings point to specific prenylation events that may be key players in AD progression. This platform can be refined for biomarker discovery and applied to drug response assessment in translational research.
References
- Distefano M.D., Albers L.N., Xu J. (2005) Protein Prenylation. In: Encyclopedic Reference of Genomics and Proteomics in Molecular Medicine. Springer. https://link.springer.com/rwe/10.1007/3-540-29623-9_2750
- Charuta C. Palsuledesai and Mark D. Distefano Protein Prenylation: Enzymes, Therapeutics, and Biotechnology Applications ACS Chemical Biology 2015 10 (1), 51-62. DOI: 10.1021/cb500791f
- Fang L. Zhang and Patrick J. Casey PROTEIN PRENYLATION: Molecular Mechanisms and Functional Consequences Annu Rev. Biochem. 1996. 65:24149. DOI: 10.1146/annurev.bi.65.070196.001325
- Jeong A, Auger SA, Maity S, Fredriksen K, Zhong R, Li L, Distefano MD. In Vivo Prenylomic Profiling in the Brain of a Transgenic Mouse Model of Alzheimer's Disease Reveals Increased Prenylation of a Key Set of Proteins. ACS Chem Biol. 2022. PMID: 36109170











