Isopentenyl Adenosine Analysis Service
Creative Proteomics provides ultra-sensitive LC-MS/MS analysis of Isopentenyl Adenosine and related metabolites, enabling accurate quantification across various sample types. Our service supports RNA modification research, biomarker discovery, and pathway analysis, helping scientists decode metabolic regulation and disease mechanisms with precision and reproducibility.
Submit Your Request Now
×- What We Provide
- Advantage
- Workflow
- Technology Platform
- Sample Requirements
- FAQ
- Publication
What is Isopentenyl Adenosine?
Isopentenyl Adenosine is a naturally occurring modified nucleoside and a biologically active derivative of adenosine. It belongs to the family of isopentenylated purines and plays a critical role in diverse cellular processes, particularly in regulating RNA modification, signal transduction, and metabolic activity. In both plant and animal systems, Isopentenyl Adenosine functions in post-transcriptional gene regulation through its involvement in tRNA modification, influencing translation fidelity and efficiency.
Due to its low abundance and structural similarity to other purine nucleosides, detecting and quantifying Isopentenyl Adenosine requires sophisticated analytical platforms. Understanding its behavior and concentration across various biological matrices is key to uncovering metabolic patterns, disease markers, and biochemical regulatory mechanisms.
Isopentenyl Adenosine Analysis Service Offered by Creative Proteomics
- Targeted Quantitative Analysis of Isopentenyl Adenosine: Absolute quantification using isotope-labeled internal standards via LC-MS/MS to ensure high sensitivity and precision.
- Detection of Isopentenyl-Related Nucleotides: Analysis of upstream and downstream metabolites such as Isopentenyl AMP, Isopentenyl ADP, and related intermediates in purine and cytokinin pathways.
- Pathway-Focused Metabolite Profiling: Integration of isopentenyl adenosine data into curated metabolic pathways, supporting systems biology studies and biomarker discovery.
- Sample Type Flexibility: Validated for a broad range of matrices including plasma, serum, tissue, cultured cells, plant extracts, and microbial samples.
- Custom Assay Development: Tailored method development and validation to meet specific experimental goals or novel matrix types.
- Batch Processing & High-Throughput Capability: Scalable service options for large cohort studies, screening projects, or multi-condition experiments.
- Data Interpretation Support: Optional bioinformatics analysis and visualization of isopentenyl adenosine-related metabolic trends and pathway impacts.
List of Detected Isopentenyl Adenosine and Related Metabolites
| Category | Analyte Name | Biological Role / Pathway |
|---|---|---|
| Primary Target | Isopentenyl Adenosine (i⁶A) | tRNA modification; translation regulation |
| Upstream Precursors | Isopentenyl Pyrophosphate (IPP) | Mevalonate pathway; cytokinin biosynthesis |
| Dimethylallyl Pyrophosphate (DMAPP) | Precursor for prenylation reactions | |
| AMP (Adenosine Monophosphate) | Purine metabolism | |
| tRNA with unmodified adenosine (A37) | Precursor to i⁶A modification | |
| Related Modified Nucleosides | 2-Methylthio-Isopentenyl Adenosine (ms²i⁶A) | Modified form in mitochondrial/archaeal tRNAs |
| N6-Isopentenyl Adenosine Monophosphate | Intermediate in enzymatic transfer pathways | |
| Byproducts / Degradation | Adenosine | Nucleoside metabolism |
| Inosine | Result of A-to-I RNA editing | |
| Hypoxanthine | Purine degradation product | |
| Associated Pathways | Cytokinin Ribosides (e.g., Isopentenyladenine Riboside) | Plant hormone signaling |
| Methylthioadenosine (MTA) | Linked to polyamine and purine salvage pathways | |
| S-Adenosylmethionine (SAM) | Methyl donor for tRNA modification |
Advantages of Isopentenyl Adenosine Assay
- Ultra-High Sensitivity: Detection limit (LOD) as low as 0.1 femtomole (fmol) using triple quadrupole LC-MS/MS, allowing accurate measurement of trace levels in complex matrices.
- Excellent Quantitative Precision: Relative standard deviation (RSD) of<5% across technical replicates; linearity (R²) of calibration curves typically exceeds 0.998 across the dynamic range.
- High Recovery Rate: Optimized extraction protocols yield ≥85% recovery of Isopentenyl Adenosine from plasma, serum, and plant matrices, minimizing sample loss.
- Wide Dynamic Range: Accurate quantification from 0.1 fmol to 1000 pmol, enabling simultaneous detection of basal and elevated metabolite concentrations.
- High Throughput Capacity: Capability to process 96–384 samples per batch, suitable for large-scale biomarker studies and population-based metabolomics.
- Cross-Matrix Compatibility: Validated workflows for multiple sample types, including human/animal biofluids, tissues, cell cultures, and plant extracts—with no loss in sensitivity or specificity.
- Data Traceability & Reproducibility: Use of internal standards and QC samples in every batch ensures analytical consistency across studies and platforms.
Workflow for Isopentenyl Adenosine Analysis Service
Technology Platform for Isopentenyl Adenosine Analysis Service
Primary Platform: LC-MS/MS using Sciex QTRAP 6500+ offers femtomole-level sensitivity and excellent reproducibility.
Chromatographic System: UHPLC system (Agilent 1290 Infinity II) equipped with C18 reversed-phase columns for sharp peak resolution.

SCIEX Triple Quad™ 6500+ (Figure from Sciex)

Agilent 1260 Infinity II HPLC (Fig from Agilent)
Sample Requirements for Isopentenyl Adenosine Analysis Service
| Sample Type | Minimum Amount Required | Notes |
|---|---|---|
| Plasma / Serum | ≥ 200 µL | Use EDTA or heparin tubes; avoid hemolysis |
| Whole Blood | ≥ 500 µL | Requires pre-processing to isolate nucleosides |
| Cell Pellet | ≥ 1 × 10⁷ cells | Washed with PBS; snap frozen or stored at −80°C |
| Animal Tissue | ≥ 50 mg | Flash-frozen preferred; homogenization required before extraction |
| Plant Tissue / Leaf | ≥ 100 mg | Fresh or flash-frozen; avoid degradation from light or heat |
| Microbial Culture Pellet | ≥ 50 mg wet weight / 1 × 10⁹ cells | Washed and frozen immediately post-centrifugation |
| Cell Culture Medium | ≥ 500 µL | Collect before media acidification; filter or centrifuge to remove cells |
| Urine | ≥ 1 mL | Midstream, fresh or frozen; no preservatives |
Applications of Isopentenyl Adenosine Assay Service
Epitranscriptomics Research
Investigating RNA modifications and their roles in gene regulation and translational control.
Plant Physiology
Studying cytokinin biosynthesis pathways and hormone-regulated growth responses.
Functional Genomics
Exploring the impact of tRNA modifications on protein expression and codon usage bias.
Metabolic Flux Analysis
Tracing purine metabolism and nucleoside turnover under varying nutrient or stress conditions.
Synthetic Biology
Engineering pathways involving prenylated nucleosides for expanded genetic coding systems.
Microbial Adaptation Studies
Analyzing RNA modification patterns in prokaryotes under environmental or antibiotic stress.
FAQ of Isopentenyl Adenosine Analysis Service
What is the recommended storage condition for my samples before shipping?
All biological samples should be stored at −80°C prior to shipment. Avoid repeated freeze–thaw cycles, as they can degrade nucleoside integrity.
Can you analyze small sample volumes or low biomass samples?
Yes, we offer low-input workflows for limited materials such as rare tissues or primary cells. Please contact us for pre-assessment of feasibility.
Do I need to prepare the samples in any specific buffer or preservative?
No preservatives are necessary. We recommend PBS washes for cell pellets, and samples should be free of detergents or phenol-based reagents.
How stable is Isopentenyl Adenosine during storage and shipping?
It is relatively stable when samples are kept frozen and protected from enzymatic degradation. We advise using dry ice shipment for optimal stability.
Can I submit different sample types in a single batch?
Yes. You may submit mixed sample types (e.g., plasma + plant tissue), though extraction and processing may differ. Please indicate the type of each sample clearly.
Is method customization available for special sample matrices or detection targets?
Yes, we offer custom method development, including matrix-specific optimization and inclusion of rare or synthetic analytes.
Can I reanalyze the same samples later for different nucleosides?
In most cases, yes. We retain extracted aliquots for up to 30 days post-analysis upon request, allowing for additional target screening.
What format will the results be delivered in?
We provide results as raw data (vendor format and .mzML), processed quantitative tables (.xlsx or .csv), and an optional PDF report with pathway integration.
Is isotope labeling necessary for quantification?
While not mandatory, stable isotope-labeled internal standards are highly recommended for accurate absolute quantification, and are included in our standard method.
Learn about other Q&A.
Isopentenyl Adenosine Analysis Service Case Study

Title: A LONELY GUY protein of Bordetella pertussis with unique features is related to oxidative stress
Journal: Scientific Reports
Published: June 9, 2021
- Background
- Materials & Methods
- Results
Bordetella pertussis, the causative agent of whooping cough, remains a significant public health concern despite widespread vaccination. The resurgence of pertussis is linked to waning immunity, strain variation, and diagnostic challenges. In the search for novel virulence factors, researchers investigated BP1253—a previously misclassified protein in B. pertussis. This study identified BP1253 as a LONELY GUY (LOG) homolog, which acts as a cytokinin-activating enzyme. Unlike typical LOGs that produce adenine-derived cytokinins, BP1253 primarily generates 6-O-methylguanine from GMP, a unique feature potentially linked to oxidative stress sensitivity in the bacterium. This finding expands the known role of LOG proteins beyond plant biology and suggests a functional role in bacterial pathogenicity.
- Materials: Reagents from Calbiochem, Bio-Rad, Roche, Pierce, Invitrogen, and Sigma Aldrich.
- Bioinformatics: Sequence analysis used BLAST and Clustal Omega; structural alignment via PDB Viewer and ESPript 3.0; visualization by PyMOL; phylogeny with MAFFT and Archaeopteryx.js.
- Bacterial Strains & Culture: B. pertussis strains BP536, BP3629, and BP36216 grown on BG agar with sheep blood, then in Stainer-Scholte medium at 37 °C.
- Recombinant Protein Expression & Purification: Synthetic bp1253 gene cloned into pET24b(+), expressed in E. coli BL21(DE3) with IPTG induction; purified via Co²⁺-affinity chromatography; LPS removed if needed.
- Size-Exclusion Chromatography: Analytical SEC using BEH200 column to assess oligomerization.
- BP1253 Knockout Generation: Allelic exchange using flanking PCR amplicons, kanamycin resistance cassette, and pSORTP1 plasmid; confirmed by PCR.
- Mutant Purification: Six point mutants expressed in E. coli; purified in 96-well Ni-sepharose plates under vacuum.
- SDS-PAGE & Western Blot: Used to assess protein purity and identity with α-BP1253 antibodies and ECL detection.
- Phosphoribohydrolase Assay: Assessed by TLC separation of nitrogenous bases after enzyme reaction.
- Mouse Immunization: BALB/c mice immunized with recombinant BP1253-His plus alum; sera collected post-immunization.
- Cytokinin Analysis (LC-MS/MS): Supernatants filtered, SPE-processed, and analyzed via LC-MS/MS with Orbitrap and UV detection.
- Surface Plasmon Resonance: BP1253 coupled to CM5 chip; nucleotide binding analyzed by Biacore T200.
- Phenotypic Characterization: Bacteria exposed to H₂O₂; survival measured to assess oxidative stress sensitivity.
- MALS Analysis: Performed post-SEC to determine molecular mass using HELEOS and ASTRA software.
- Protein Quantification: Protein concentrations determined by BCA assay using BSA standard.
BpLOG Identification: BP1253 shares conserved LOG motifs (PGGxGTxxE, catalytic Arg and Glu) and aligns phylogenetically between type-I and type-II LOGs; SEC-MALS confirmed it forms dimers.
Nucleotide Binding: BP1253 binds AMP (KD = 5.7 µM), GMP (38.6 µM), and IMP (71.8 µM); weakly binds TMP and UMP; no CMP binding detected via SPR.
Enzymatic Activity: Exhibits phosphoribohydrolase (PRH) activity, cleaving AMP, GMP, and CMP; shows stronger activity toward GMP. Site-directed mutagenesis confirmed key residues (R120, E143, K121) essential for activity.
Cytokinin Production: LC-MS/MS detected isopentenyl adenine and kinetin in both wild-type and Δ1253 lysates. A unique compound, 6-O-methylguanine, was found only in wild-type supernatants and matched synthetic standard by retention time and fragmentation pattern.
Oxidative Stress Sensitivity: Wild-type Bp showed increased sensitivity to H₂O₂ compared to Δ1253. Addition of 6-O-methylguanine restored sensitivity in Δ1253 under oxidative stress, indicating BpLOG contributes to ROS susceptibility.
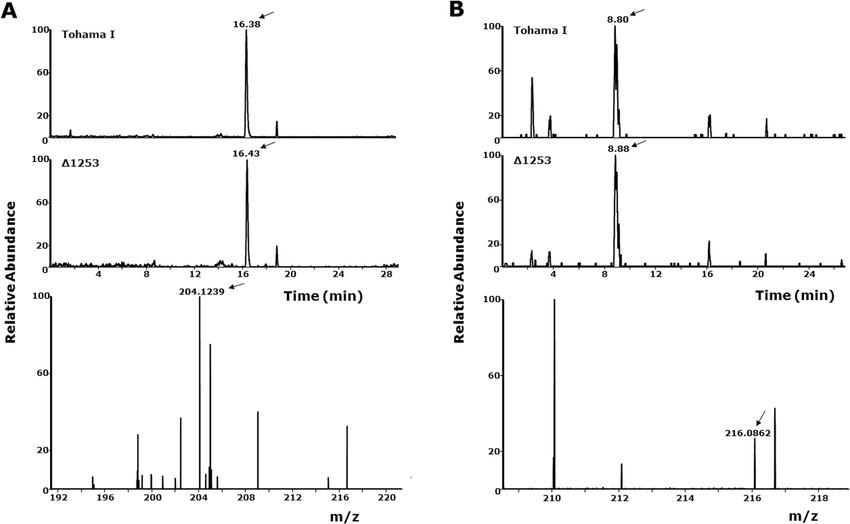 LC-MS analysis of isopentenyl adenine and kinetin in Tohama I and Δ1253 mutant lysates. (A) Extracted ion chromatogram showing isopentenyl adenine at RT 16.4 min (m/z 204.24, C₁₀H₁₄N₅, −2.21 ppm). (B) Extracted ion chromatogram showing kinetin at RT 8.8 min (m/z 216.08, C₁₀H₁₀ON₅, −8.45 ppm). Samples analyzed by positive-mode ESI-LC-MS; collision energy: 26 a.u. Data shown are representative of triplicate experiments.
LC-MS analysis of isopentenyl adenine and kinetin in Tohama I and Δ1253 mutant lysates. (A) Extracted ion chromatogram showing isopentenyl adenine at RT 16.4 min (m/z 204.24, C₁₀H₁₄N₅, −2.21 ppm). (B) Extracted ion chromatogram showing kinetin at RT 8.8 min (m/z 216.08, C₁₀H₁₀ON₅, −8.45 ppm). Samples analyzed by positive-mode ESI-LC-MS; collision energy: 26 a.u. Data shown are representative of triplicate experiments.
Reference
- Moramarco, Filippo, et al. "A LONELY GUY protein of Bordetella pertussis with unique features is related to oxidative stress." Scientific reports 9.1 (2019): 17016. https://doi.org/10.1038/s41598-019-53171-9
Publications
Here are some publications in Metabolomics research from our clients:

- Bacterial–fungal interactions revealed by genome-wide analysis of bacterial mutant fitness. 2021. https://doi.org/10.1038/s41564-020-00800-z
- Non-invasive elevation of circulating corticosterone increases the rejection of foreign eggs in female American robins (Turdus migratorius). 2022. https://doi.org/10.1016/j.yhbeh.2022.105278
- Characterization of CYCLOPHILLIN38 shows that a photosynthesis-derived systemic signal controls lateral root emergence. 2021. https://doi.org/10.1093/plphys/kiaa032
- Inflammation primes the kidney for recovery by activating AZIN1 A-to-I editing. 2023. https://doi.org/10.1101/2023.11.09.566426
- A personalized probabilistic approach to ovarian cancer diagnostics. 2024. https://doi.org/10.1016/j.ygyno.2023.12.030






