4D-DIA Quantitative Proteomics Service
Cohort-ready 4D-DIA proteomics (TIMS + DIA)
We provide deep, reproducible quantification using 4D proteomics with ion mobility (TIMS) and DIA acquisition. Our service is built for cohorts (30–200+ samples) with batch-aware QC and decision-grade reporting, ensuring fewer missing values and stronger consistency compared to traditional DDA workflows.
Key Highlights:
- Bruker timsTOF HT (TIMS-XR) for true 4D separation (RT, m/z, intensity, mobility)
- Acquisition flexibility: PASEF® / dia-PASEF® / prm-PASEF®
- CCS-assisted identification to improve confidence and cross-run consistency
- Optional CPDB3000™ deep plasma/serum workflow for low-abundance biomarkers
Request a Study Design Plan
×
- Sample types: tissue | cells | plasma/serum | CSF | microbes | plants
- Deliverables: quant matrix + QC report (optional differential/pathway modules)
- Use cases: biomarker | MoA/PD | immunology/inflammation | cohort proteomics CRO
- RUO: Research Use Only
- Overview
- Platform
- Workflow
- Deep Blood
- Samples
- Proof & Quality
- Bioinformatics
- Packages
- Case Study
- FAQs
Understanding 4D-DIA Proteomics
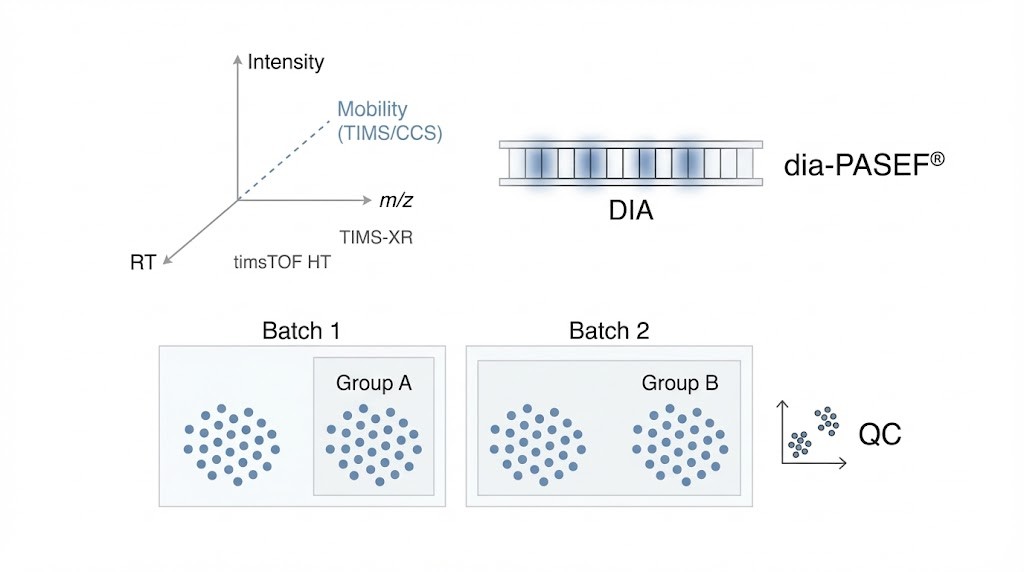
Traditional LC-MS/MS proteomics separates peptides in three dimensions: retention time (RT), mass-to-charge ratio (m/z), and signal intensity. 4D proteomics introduces a fourth, orthogonal separation dimension: ion mobility.
The Power of Ion Mobility (TIMS)
- Ion mobility separates ions in the gas phase based on their size, shape, and charge, generating a collisional cross section (CCS) value.
- This extra dimension reduces spectral complexity before ions even reach the mass analyzer.
- When combined with data-independent acquisition (DIA), it enables more complete peptide sampling and improved discrimination of co-eluting species.
Why it matters for cohorts
In practice, 4D-DIA means fewer missing values, stronger quantitative consistency, and better scalability to large cohorts compared with DDA-based workflows.
Platform & Acquisition Modes
Our service is powered by the timsTOF HT mass spectrometry platform, which integrates TIMS-XR (Trapped Ion Mobility Spectrometry) with fast TOF detection.
Addressing Complexity
- In complex samples like plasma or tissues, many peptides co-elute. Without sufficient separation, this leads to fragment ion interference.
- TIMS provides an additional separation step before fragmentation, significantly reducing these issues and stabilizing quantification across runs.
Supported Acquisition Strategies
- PASEF®: Maximizes sensitivity by synchronizing ion accumulation and fragmentation.
- dia-PASEF®: Combines DIA with ion mobility, increasing ion utilization efficiency and coverage.
- prm-PASEF®: Enables targeted follow-up for hypothesis-driven validation studies.
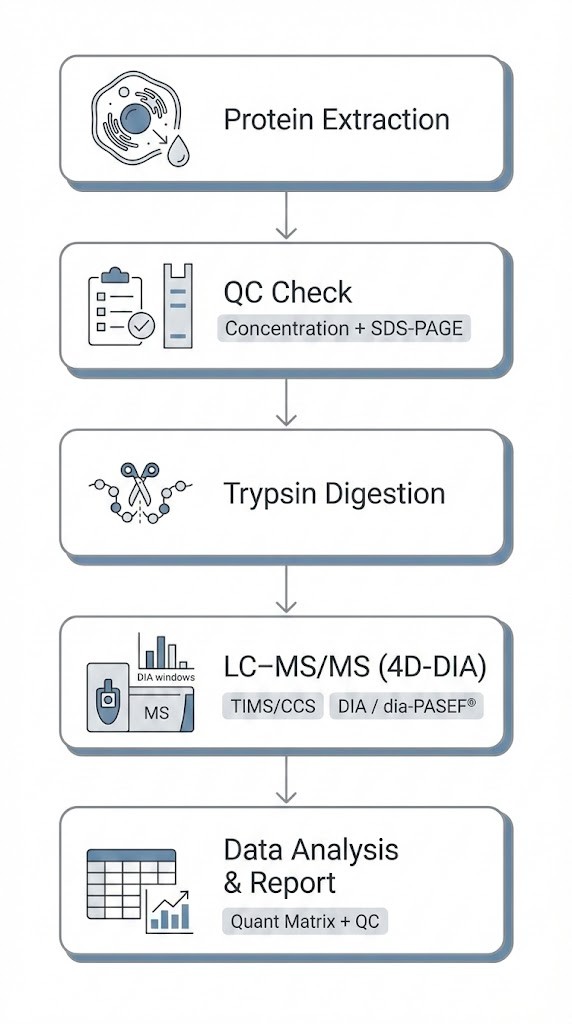
Experimental Workflow (Wet Lab)
A high-quality proteomics dataset begins long before mass spectrometry. Our workflow emphasizes standardization and quality control at every step.
1. Protein Extraction & QC
Optimized protocols are applied based on tissue type, cell composition, or biofluid matrix. We perform protein concentration measurement and SDS-PAGE inspection to confirm integrity.
2. Enzymatic Digestion
Proteins are digested into peptides under controlled conditions to support reproducibility.
3. LC–MS/MS Acquisition (4D-DIA)
DIA-based acquisition is performed with ion mobility separation (TIMS/CCS) using dia-PASEF® modes tailored to the project.
4. Data Processing
Internal controls are applied to minimize technical variability, followed by bioinformatics analysis.
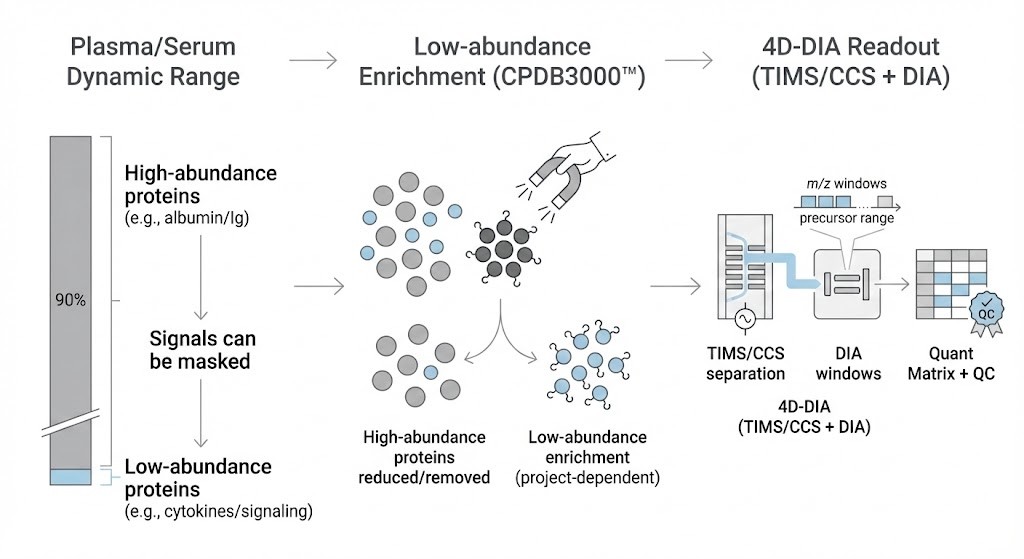
Deep Blood / Plasma Option (CPDB3000™)
For plasma/serum studies where low-abundance coverage is a priority, we offer CPDB3000™ (Deep Blood Proteome by 4D-DIA). This workflow addresses the dynamic range challenge of blood proteomics.
Workflow Features
- Low-abundance enrichment: Uses magnetic nano-material–based capture (project-dependent) to reduce high-abundance interference (e.g., albumin).
- 4D-DIA acquisition: Uses TIMS + DIA for improved sensitivity and consistency.
- Curated database strategy: Supports confident identification and interpretable annotation.
Sample Requirements
This service supports a broad spectrum of biological materials.
| Sample Type | Typical Input |
|---|---|
| Animal or human tissues | 10–20 mg |
| Cultured or primary cells | 1–5 × 10⁶ cells |
| Plasma / serum / CSF / other biofluids | 50–200 µL |
| Plant tissues | 100 mg–5 g |
| Microbial pellets | 100–500 mg |
| Culture supernatant | 20–30 mL |
| Protein solutions | 30–50 µg |
Note: If sample quantity or quality is a concern, pre-submission consultation is strongly recommended.
Proof & Quality
High-quality proteomics is defined by repeatable performance across samples, batches, and time. Our 4D-DIA service is built around standardized SOPs and batch-aware QC.
Quality System
| Phase | Checks Performed |
|---|---|
| Pre-analytical | Protein quantity check, SDS-PAGE integrity review, sample acceptance review. |
| Run-level (Acquisition) | Instrument performance tracking, RT alignment checks, QC sample monitoring. |
| Bioinformatics QA | FDR-controlled reporting, batch-aware summaries (PCA, CV), traceability documentation. |
Data Analysis & Bioinformatics
High-quality proteomics data must be interpretable, traceable, and publication-ready. Our bioinformatics workflow ensures this through rigorous statistical control.
Standard Analysis
- Data Organization: Peptide/protein identification with FDR control and quantitative matrix generation.
- Quality Metrics: PCA for sample clustering, expression distribution profiles, and variability indicators (RSD/CV).
Optional Advanced Analysis
- Differential Expression: Statistical testing, volcano plots, and hierarchical clustering heatmaps.
- Functional Annotation: Mapping to GO, KEGG, InterPro, Reactome, and WikiPathways.
Use Cases Aligned with ICP
- Biomarker Discovery (N: 30–200+): 4D-DIA provides consistent detection across cohorts, reducing missing values that obscure true signals.
- Drug MoA & PD Studies: Ideal for multi-dose/time-course designs to identify pathway-level responses.
- Immuno-Oncology: Stable quantification of immune-related pathways in heterogeneous samples.
- CRO & Platform-Scale: Emphasis on SOP stability and reproducibility for large-scale delivery.
Platform Comparison
| Feature | 4D-DIA (timsTOF HT) | Astral DIA |
|---|---|---|
| Separation | Orthogonal 4D (TIMS ion mobility) | High-speed scanning |
| Strength | Reduced interference, strong in complex/low-abundance samples | Maximum throughput on short gradients |
| Recommendation | Choose for complexity, interference control, and deep plasma | Choose for extreme throughput |
Packages & Pricing Logic
We offer common scope options that can be mixed and matched. Final scope is confirmed during study design review.
| Scope Option | Goal | Inclusions |
|---|---|---|
| A: Feasibility / Pilot | Confirm sample suitability | Standard QC + 4D-DIA acquisition, Quant matrix + QC summary. |
| B: Cohort Quantification | Cohort-ready quantification | Batch-aware planning, Full matrix + cohort QC report (PCA, CV), Optional differential analysis. |
| C: Cohort + Insights | Decision-grade reporting | Cohort quantification + biological interpretation, Pathway prioritization, Publication figures. |
Example Deliverables
Typical deliverables include quantitative results tables, QC summary reports, and optional differential/enrichment packages.
Case Study: MultiPro Benchmark
Wang H, Lim KP, Kong W, et al. MultiPro: DDA-PASEF and diaPASEF acquired cell line proteomic datasets with deliberate batch effects.Scientific Data (2023) 10:858. (CC BY 4.0)
https://www.nature.com/articles/s41597-023-02779-8
- Background
- Methods
- Results
- Conclusions
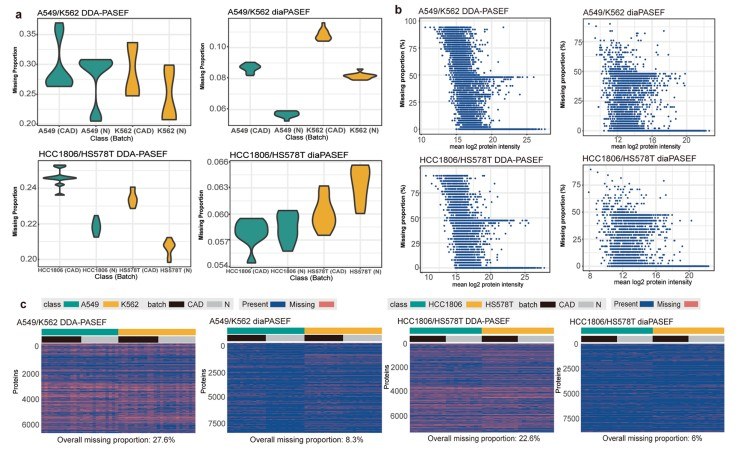
Cohort-scale proteomics often fails due to technical issues—especially missing values and batch effects. To support benchmarking, the MultiPro team built large-scale proteomics datasets with deliberate batch effects, including DIA-mode diaPASEF data on timsTOF instruments.
The study used well-characterized human cell line pairs (A549 vs K562) and designed datasets with biological replicates and technical replicates. Samples were intentionally run on two timsTOF Pro instruments in parallel to introduce batch structure. The A549/K562 diaPASEF datasets contained 48 samples split into two batches.
Depth: MultiPro reported ~7,994–8,282 protein groups per run on average, and ~8,800+ total.
Reproducibility: diaPASEF datasets showed very high replicate correlations (Spearman > 0.99) and low CVs.
Missing Values: Overall missingness dropped from >20% in DDA mode to ~6% in DIA mode, demonstrating superior data completeness for cohorts.
This benchmark highlights that cohort-ready DIA data can meaningfully reduce missingness and support consistent quantification. It emphasizes the importance of standardized SOPs and batch-aware QC (like PCA/PVCA) to manage technical variance in large-scale studies.
FAQs
What is 4D-DIA proteomics?
4D-DIA combines ion mobility (TIMS) with DIA to reduce interference and support stable quantification. It adds a fourth dimension (CCS) to standard LC-MS/MS separation.
When should a team choose 4D-DIA instead of DDA?
When you need consistent quantification across many samples and want fewer missing values than DDA/top-N workflows. It is ideal for cohorts where data completeness is critical.
Do you support dia-PASEF®?
Yes. dia-PASEF integrates DIA with TIMS separation and can improve sensitivity and coverage in complex samples. We also support prm-PASEF for targeted validation.
What projects are the best fit for 4D-DIA?
- Biomarker discovery/verification in plasma/serum cohorts.
- Drug Mechanism-of-Action & PD Studies.
- Immuno-Oncology & Inflammation Research requiring stable quantification.
What sample types do you accept?
We accept tissues, cells, biofluids (plasma/serum/CSF), microbes, plants, culture supernatants, and protein solutions.
What deliverables will I receive?
You will receive a quantitative matrix, QC report, summary figures, and optional differential/pathway reports. Raw and processed files are also provided for transparency.
How do you evaluate reproducibility and missing values?
We provide PCA, distribution summaries, and variability indicators (e.g., RSD/CV) in our QC reports to evaluate stability across batches.
Can you handle multi-batch or longitudinal studies?
Yes. We plan batch structure up front and provide QC summaries to evaluate stability across batches, ensuring data is comparable throughout the study.
Do you provide differential expression and pathway analysis?
Yes. Options include differential analysis, GO/KEGG enrichment, and pathway databases (e.g., Reactome/WikiPathways for human/mouse) to generate biological insights.
Learn about other Q&A .
References
- Bruker. timsTOF HT (TIMS-XR, CCS-enabled). Accessed 2026-01-28.
- Bruker. PASEF® acquisition modes. Accessed 2026-01-28.
- Meier F, et al. diaPASEF: parallel accumulation–serial fragmentation combined with data-independent acquisition. Nature Methods. 2020.
- Demichev V, et al. DIA-NN: neural networks and interference correction enable deep proteome coverage. Nature Methods. 2020.
- Röst HL, et al. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nature Biotechnology. 2014.
- Wang H, et al. MultiPro: DDA-PASEF and diaPASEF acquired cell line proteomic datasets with deliberate batch effects. Scientific Data. 2023.
- The UniProt Consortium. UniProt: the Universal Protein Knowledgebase in 2025. Nucleic Acids Research. 2025.





