MS-based Spatial Proteomics Service (LCM/LMD-guided LC–MS/MS)
Spatially Resolved Proteomics for Tissue Microenvironments, Biomarker Discovery, and Mechanism Studies (RUO)
See protein biology in the right place.
Our MS-based spatial proteomics service measures proteins from specific regions of a tissue section—also called regions of interest (ROIs)—using laser microdissection (LCM/LMD) followed by high-sensitivity LC–MS/MS. You get region-by-region protein and pathway changes, with clear links back to histology and morphology.
Best fit for:
- Tumor microenvironment (TME) niches and resistance regions
- Pathology-guided region comparisons (margin vs core, invasive front, necrosis, stroma, immune infiltrates)
- Archival cohorts that are hard to profile with bulk proteomics (including FFPE)
What you receive (not just a matrix):
- ROI-level protein quantification + QC
- Differential proteins across ROIs / phenotypes
- Pathway and network interpretation (a clear biological story)
- A reproducible analysis package (methods, parameters, versions)
Choose your workflow (no proprietary platform lock-in):
Submit Your Request Now
×
- Overview
- Applications
- Workflows
- Process
- Deliverables
- Methods
- QC
- Samples
- Turnaround
- FAQ
What is MS-based Spatial Proteomics?
MS-based spatial proteomics (also called spatially resolved proteomics) measures proteins from defined tissue microregions while keeping the location context. ROIs are selected on histology, isolated by LCM/LMD, and quantified by LC–MS/MS.
In one sentence: you get ROI-by-ROI protein and pathway changes, linked to tissue morphology.
When should you use it?
Choose this approach when:
- You need deep protein profiling from defined microregions (tumor core vs margin, stroma vs immune infiltrates).
- Your samples are heterogeneous, and bulk proteomics would mix signals and hide local biology.
- You want a workflow aligned with peer-reviewed MS-based tissue protocols, without dependence on proprietary spatial chemistries.
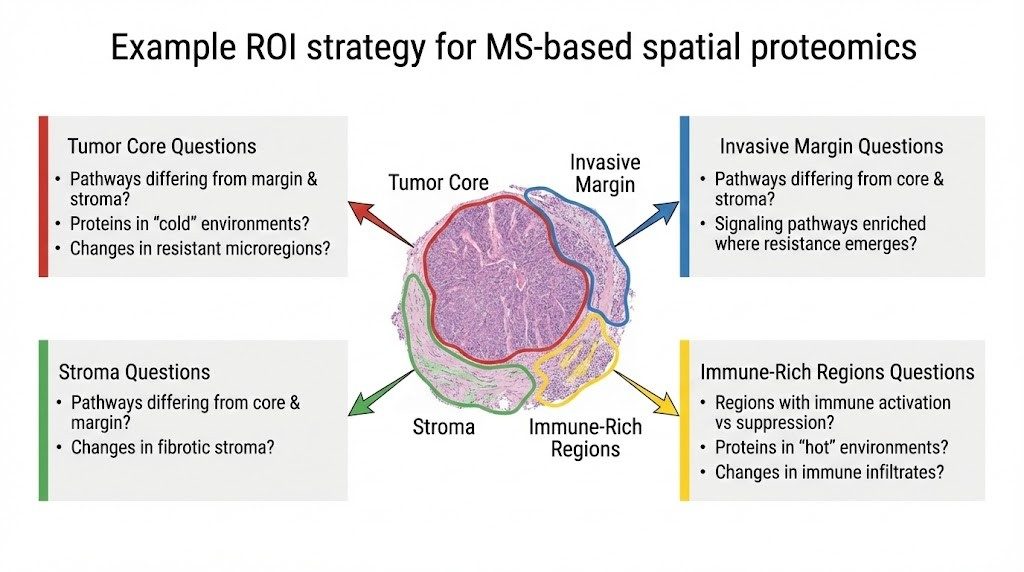
What Questions Can We Answer?
Tumor microenvironment (TME) niches
- Which regions show immune activation vs suppression?
- Which pathways differ between the tumor core, the invasive margin, and the stroma?
- Which proteins track with "cold" vs "hot" immune microenvironments?
Mechanism and resistance
- What changes appear in resistant microregions (hypoxic niches, fibrotic stroma, invasive fronts)?
- Which signaling pathways are enriched where resistance emerges?
Biomarker discovery and cohort stratification (RUO)
- Can we build ROI-based protein signatures linked to histology features?
- Can we compare responder vs non-responder patterns by matching ROIs across samples?
Non-oncology tissues (inflammation, fibrosis, neuro, infection)
- Region-specific disease pathways (lesion vs adjacent normal)
- Microanatomy-driven protein programs
Spatial proteomics is increasingly used in translational research because tissue biology is spatially organized—especially in complex microenvironments.
Core Workflows We Offer
1) LCM/LMD-Guided Spatial Proteomics (Core Service)
Best for: deep, region-by-region protein biology and pathway interpretation.
How it works (high level):
- Histology review & ROI plan (H&E and/or IF guidance)
- Laser microdissection (LCM/LMD) to capture target regions
- Low-input protein extraction and digestion
- LC–MS/MS measurement (DDA or DIA, depending on study goals)
- ROI-level statistics + pathway interpretation
This approach is a common foundation for spatially resolved proteome mapping in peer-reviewed studies.
2) FFPE Spatial Proteomics (Archival Cohorts)
Best for: retrospective cohorts and biobanked tissue.
FFPE proteomics has matured rapidly, with protocols designed to handle de-crosslinking and detergent removal so quantitative LC–MS/MS can be applied to limited FFPE material.
Published high-throughput FFPE workflows can achieve deep proteome coverage—in some studies reporting ~8,000–10,000 proteins per sample under optimized conditions—while maintaining reproducibility suitable for research cohort design.
We do not promise a fixed number, because tissue type, age, fixation, and ROI size all matter. Instead, we design the project to maximize interpretability and reproducibility.
3) MALDI-MSI Add-on (Whole-Slide Molecular Maps)
Best for: visualizing spatial distributions across the tissue and guiding ROI selection.
MALDI mass spectrometry imaging (MALDI-MSI) maps molecular signals directly in tissue sections and has a deep review literature describing tissue applications, strengths, and practical limits.
Recent reviews also describe MSI-guided spatial proteomics workflows where MSI helps identify ROIs that are then interrogated by chromatography-based proteomics methods.
How to use it in practice:
- Start with MALDI-MSI to see where signals concentrate.
- Then use LCM/LMD to isolate those ROIs for deep LC–MS/MS protein identification and pathway analysis.
 An open MS-based spatial proteomics workflow reduces platform lock-in and aligns with widely published protocols (RUO).
An open MS-based spatial proteomics workflow reduces platform lock-in and aligns with widely published protocols (RUO).
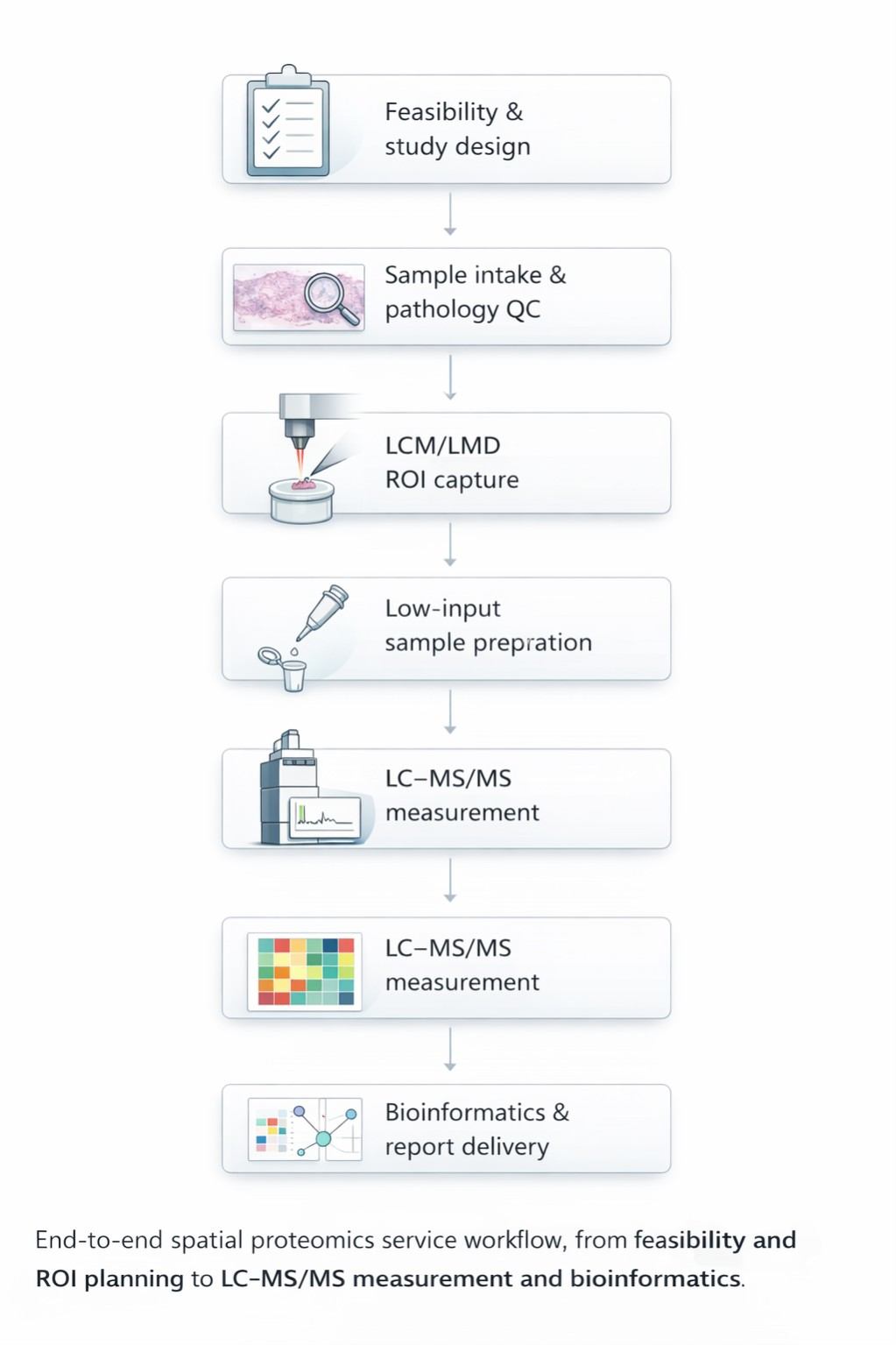
End-to-End Workflow (What Happens After You Contact Us)
Step 1 — Feasibility & Study Design (Scientist-to-scientist)
We align on:
- Tissue type and condition (FFPE vs frozen)
- ROI strategy (how many regions, labels, matching across cases)
- Primary goal: discovery, mechanism, cohort comparison, or method validation
Output: a short study plan with sample requirements and ROI guidance.
Step 2 — Sample Intake & Pathology QC
We confirm:
- Section thickness, slide type, storage conditions
- Whether H&E/IF staining will be used for ROI selection
- Whether tissue integrity supports clean ROI boundaries
Why this matters: Spatial proteomics is only as good as the ROI definition. LCM was created to isolate specific cells/regions in heterogeneous tissue under direct visualization.
Step 3 — ROI Capture by LCM/LMD
We capture the agreed ROIs. For cohort work, we standardize ROI size and region definitions to reduce "apples-to-oranges" comparisons.
Step 4 — Low-Input Sample Prep (Built for Microregions)
Low-input microdissection samples require careful handling to avoid losses. Peer-reviewed methods (such as nanoPOTS coupled with LCM) were developed to improve sensitivity for laser-captured tissue samples.
Step 5 — LC–MS/MS Measurement
We choose acquisition based on your goal:
- DDA for discovery and library building
- DIA for consistent quantification across many ROIs
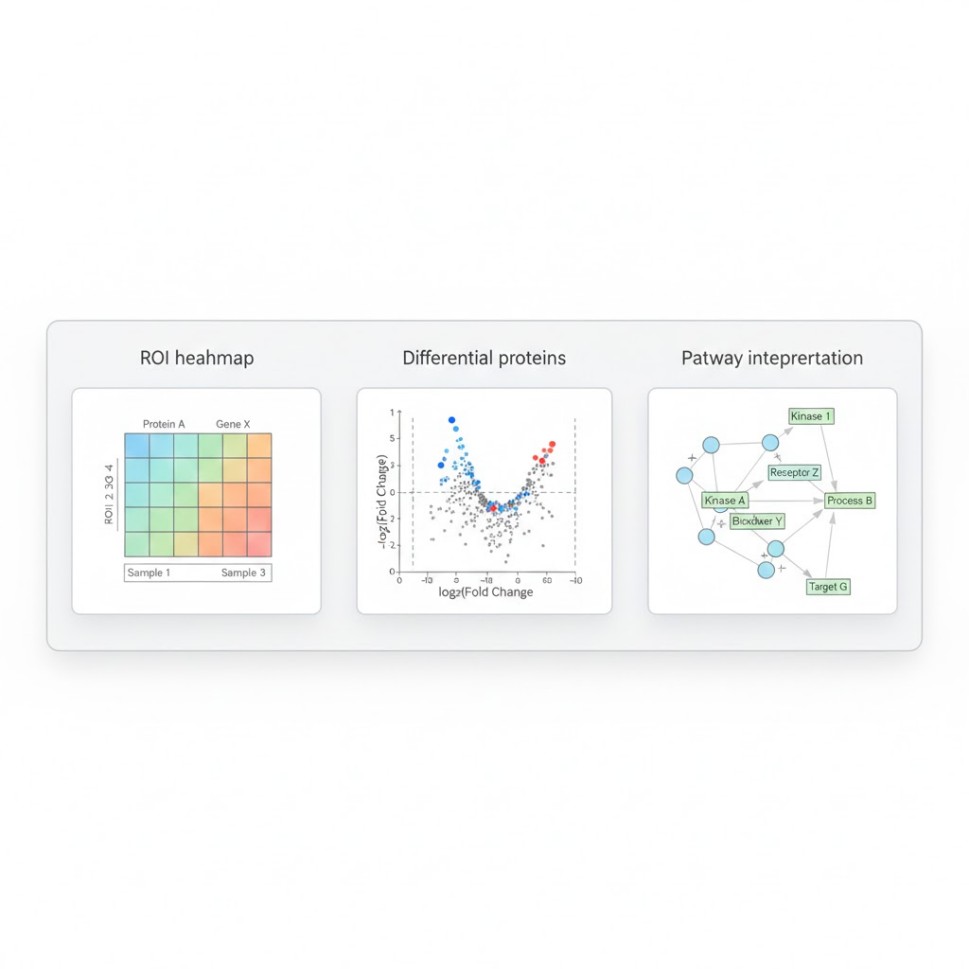
Step 6 — Analysis & Interpretation (Where the value is created)
You receive:
- ROI-level protein matrix with QC annotations
- Differential proteins (region vs region, group vs group)
- Pathway and network summaries
- Visual outputs linked back to ROIs (heatmaps, volcano plots, pathway views)
Deliverables
Data package (reproducible and audit-friendly)
- Raw files/identification outputs (as applicable)
- ROI-level quantitative matrix
- QC summary (missingness, intensity distributions, replicate behavior)
- Sample and ROI metadata (what each ROI represents)
Interpretation package (the "so what")
- Differential protein lists per comparison
- Pathway enrichment and biological themes
- Short, plain-English executive summary
- Figures ready for slide decks and manuscripts
Optional add-ons
- MALDI-MSI ion images and co-registered overlays (where feasible)
- Phosphoproteomics for signaling-centric questions (availability depends on design; deep FFPE phosphoproteome coverage has been reported in optimized workflows).
Protocol-aligned Methods
Our workflow is designed around peer-reviewed, widely used approaches in MS-based spatial tissue proteomics:
- FFPE protocol alignment: We follow key concepts described in a Nature Protocols workflow for spatially resolved analysis of FFPE tissue proteomes by quantitative mass spectrometry (FFPE processing and ROI-linked comparisons).
- LCM best-practice alignment: ROI isolation is guided by guidance summarized in a recent Nature Primer on laser capture microdissection, including how and when to use LCM in modern spatial studies.
- Low-input sensitivity strategies: For microdissected ROIs, we adopt sensitivity-first handling principles consistent with LCM-to-nanoPOTS spatial proteomics workflows.
- MALDI-MSI add-on grounding: Our MSI option is based on established MALDI imaging literature and recent discussions of MSI-guided spatial proteomics.
Quality Control and Reproducibility
We build trust the same way strong labs do: by showing our work.
QC approach (typical):
| QC Component | Description |
|---|---|
| Contamination controls | Blanks and internal standards (as needed) |
| ROI consistency checks | Region definition, cohort harmonization |
| Data metrics | Data distribution and missingness tracking |
| Reporting | Clear reporting of software, versions, and key parameters |
Why we emphasize this: spatial studies can fail silently if ROIs are inconsistent or batch effects dominate. Published frameworks explicitly focus on scalability and low-input robustness for spatial tissue proteomics.
Sample Types We Commonly Support
| Sample Type | Characteristics | Notes |
|---|---|---|
| FFPE (Formalin-Fixed, Paraffin-Embedded) | Strong for retrospective cohorts and pathology-defined ROIs | Requires de-crosslink-aware preparation; established protocols exist. |
| Fresh frozen tissue | Often higher molecular integrity | Good for certain sensitive pathway questions (design dependent). |
| Tissue microarrays (TMA) | Useful for standardized cohort comparisons | Case-by-case feasibility. |
Note: We do not overpromise. Tissue age, fixation conditions, ROI size, and biological complexity influence identifications and quantitative stability.
Spatial proteomics has strong translational potential, but our offering is Research Use Only (RUO).
Turnaround Time (What Drives It)
We provide a project-specific timeline after feasibility review. Typical drivers:
- Number of samples and ROIs
- FFPE vs frozen
- Depth of analysis (discovery vs targeted follow-up)
- Add-on imaging (MALDI-MSI)
Request a Quote (Fastest Path)
To scope your project quickly, send:
- Sample type (FFPE or frozen), tissue type, species
- Number of cases and expected number of ROIs per case
- ROI definition (pathology regions, markers, or phenotype labels)
- Main comparisons (margin vs core; group A vs group B)
- Whether you want the MALDI-MSI add-on mapping
- Any constraints (timeline, data format, downstream integration)
Frequently Asked Questions (FAQ)
1) What is the minimum ROI size you can work with?
It depends on tissue density, preparation method, and depth targets. We recommend an ROI strategy during the feasibility review. Many published spatial MS workflows are designed for ultra-low-input samples, but practical thresholds must be tailored to your tissue and question.
2) Can you do FFPE spatial proteomics for older archived blocks?
Often yes, but performance varies with fixation conditions, storage time, and ROI size. We can assess risk with a pilot design when needed.
3) Do you provide guidance for ROI selection?
We can work with your pathology annotations or collaborate on ROI rules so regions are consistent across samples.
4) Do I need immunostaining?
Not always. H&E-guided ROI selection is common. IF can be helpful when ROIs are defined by marker-positive regions.
5) What is the difference between LCM/LMD + LC–MS/MS and MALDI-MSI?
LCM/LMD + LC–MS/MS gives deeper protein identification from selected regions. MALDI-MSI gives whole-slide distribution maps and can guide ROI selection.
6) Will you share analysis methods and parameters?
Yes. We provide a reproducibility-oriented summary (software, versions, and key parameters) with the report.
7) Is this service for clinical diagnosis?
No. This service is Research Use Only (RUO) and is not intended for diagnostic procedures.
8) Can you support cohort-scale projects?
Yes. Cohort design depends on standardizing ROI rules and controlling batch effects. Optimized high-throughput FFPE workflows have been published for research-scale projects.
9) What do you deliver besides the data?
A structured, decision-ready package: QC + ROI-level results + interpretation + figures, plus an optional review call.
10) Do you support confidential projects and NDAs?
Yes (standard practice). We can align on secure transfer and data retention during scoping.
Learn about other Q&A.
References
- Springer Nature Primer — Laser capture microdissection. Laser capture microdissection. Nature Reviews Methods Primers, 2025. DOI: https://doi.org/10.1038/s43586-025-00453-4.
- Nature Protocols — Spatially resolved FFPE proteomics by quantitative MS. Spatially resolved analysis of FFPE tissue proteomes by quantitative mass spectrometry. Nature Protocols, 2020. DOI: https://doi.org/10.1038/s41596-020-0356-y Spatially resolved proteome mapping via nanoPOTS + LCM. Spatially resolved proteome mapping of laser capture microdissected tissue by nanoPOTS-based proteomics. Molecular & Cellular Proteomics, 2019. DOI: https://doi.org/10.1074/mcp.TIR118.000686
- Ultra-low-input spatial tissue proteomics framework (imaging + microdissection + MS). A framework for ultra-low-input spatial tissue proteomics. Cell Systems, 2023. DOI: https://doi.org/10.1016/j.cels.2023.07.004
- High-throughput FFPE deep proteome & phosphoproteome (8,000–10,000 proteins). High-throughput and deep proteomic and phosphoproteomic profiling of FFPE tissues. Molecular & Cellular Proteomics, 2025. DOI: https://doi.org/10.1016/j.mcpro.2025.100764
- Review — Spatial proteomics in translational and clinical research. Spatial proteomics in translational and clinical research. Nature Reviews Clinical Oncology, 2025. DOI: https://doi.org/10.1038/s44320-025-00101-9
- Review — MALDI imaging mass spectrometry: frontiers and tissue applications. MALDI imaging mass spectrometry: current frontiers and applications in tissue analysis. Laboratory Investigation, 2015. DOI: https://doi.org/10.1038/labinvest.2014.156
- Review — MSI-guided spatial proteomics (MSI → ROI → LC-MS).MALDI mass spectrometry imaging–guided spatial proteomics workflows. Critical Reviews in Analytical Chemistry, 2025. DOI: https://doi.org/10.1080/14789450.2025.2537212





