- Service Details
- Case Study
What are Low Molecular Weight Sugars?
Carbohydrates are the most abundant and diverse components of many foods such as cereals, vegetables, fruits, potatoes, and flour. Chemically they are composed of carbon, hydrogen, and oxygen and symbolized with the chemical formula (CH2O)n. Carbohydrates can exist as individual molecules or physically associated or chemically attached to other molecules. According to the number of monomers, individual carbohydrate molecules can be classified as monosaccharides, oligosaccharides or polysaccharides. When carbohydrate molecules covalently attached to proteins, the proteins are regarded as glycoproteins. When carbohydrate molecules covalently attached to lipids, the lipids are known as glycolipids. Some carbohydrates are digestible and act as crucial energy source for humans while other carbohydrates are indigestible and fail to act as energy source. Carbohydrates that can be digested, such as lignin, are regarded as dietary fiber. It is shown that consumption of sufficient quantities of dietary fiber is of great benefits of human nutrition and health, reducing the risk of diabetes and constipation, coronary heart disease and certain types of cancers. Besides acting as an important source of energy and dietary fiber, carbohydrates also contribute to the appearance, textural and sweetness of many foods.
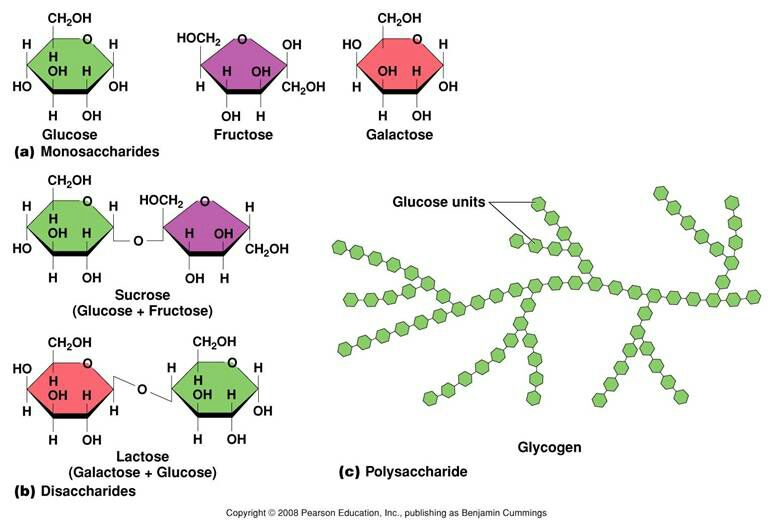
Monosaccharide, most often known simple sugar or low molecular weight sugar, is water-soluble crystalline compounds with the simplest form of carbohydrate. They are aliphatic aldehydes or ketones with one or more hydroxyl groups and one carbonyl group as reactive centers. Commonly occurring hexoses in foods are glucose, fructose and galactose, whilst commonly occurring pentose like arabinose and xylose are the most abundant monosaccharide in food. Other monosaccharide in food included glucose, fructose and galactose, which act as energy source for organisms. Monosaccharides are also building blocks for more complex carbohydrates and other macromolecules important for organisms. For example, ribose and deoxyribose are monosaccharides are the building blocks of RNA and DNA, two important genetic macromolecules crucial for life. What's more, derivatives of monosaccharides also play important roles. For example, vitamin C is a monosaccharide derivative serving as an important antioxidant.
Creative Proteomics offers a state-of-the-art analytical service for the identification and quantification of low molecular weight sugars using advanced UPLC-MS/MS (MRM) technology. This service is designed to provide high sensitivity and reproducibility, ensuring precise and accurate results for your research or industrial needs.
List of Detectable Low Molecular Weight Sugars
| Low Molecular Weight Sugars Quantified in This Service | ||
|---|---|---|
| Arabinose | Cellobiose | Fructose |
| Fucose | Galactose | Glucose |
| Lactose | Maltose | Mannose |
| Rhamnose | Ribose | Ribulose |
| Xylose | Xylulose | |
Analytical Techniques for Low Molecular Weight Sugars Analysis
At Creative Proteomics, we utilize ultra performance liquid chromatography coupled with tandem mass spectrometry (UPLC-MS/MS) in Multiple-Reaction Monitoring (MRM) mode to analyze low molecular weight sugars. This cutting-edge technique provides several critical advantages that enhance the reliability and accuracy of our analyses:
- High Sensitivity: Our UPLC-MS/MS system can detect sugar concentrations as low as 1 picogram (pg). This high sensitivity is essential for detecting low-abundance monosaccharides in complex biological samples, ensuring that even minute quantities are accurately quantified.
- Wide Dynamic Range: The method provides a linear detection range spanning three orders of magnitude (from pg to ng). This broad dynamic range allows for the accurate quantification of monosaccharides across various concentrations, making it suitable for both trace-level analysis and high-abundance measurements.
- Precision and Reproducibility: The UPLC-MS/MS (MRM) technique offers exceptional precision and reproducibility, which are crucial for generating consistent and reliable data. This level of accuracy ensures that repeated analyses yield similar results, supporting robust and dependable conclusions in research and industrial applications.
- No Need for Chemical Modifications: Unlike some other analytical methods, our approach does not require reduction, derivatization, or post-column addition of reagents. This simplifies the sample preparation process, reduces potential sources of error, and speeds up the overall analysis time.
- Multiple-Reaction Monitoring (MRM): MRM mode enhances specificity by monitoring specific precursor-product ion transitions for each analyte. This targeted approach reduces background noise and improves the detection of specific sugars in complex mixtures.
Sample Requirements
For the analysis of low molecular weight sugars, sample requirements can vary depending on the specific analytical method and instrument sensitivity. Here's a general guideline for sample types and suggested sample amounts:
| Sample Type | Sample Amount Required |
|---|---|
| Liquid samples (e.g., juices, extracts) | 1-5 mL, depending on sugar concentration |
| Solid samples (e.g., fruits, powders) | 1-5 g, finely ground or homogenized |
| Serum/plasma | 0.5-1 mL |
| Urine | 1-5 mL |
| Tissue | 20-50 mg, finely ground or homogenized |
Delivery
Upon completion of the analysis, Creative Proteomics provides a comprehensive technical report designed to give you a thorough understanding of the results obtained. The report includes:
Experiment Procedure: Detailed description of the methodologies and protocols used during the analysis.
MS/MS Instrument Parameters: Specific settings and configurations utilized on the UPLC-MS/MS system.
Analyte Concentrations: Quantitative results reported in micromolar (µM) or micrograms per milligram (µg/mg) for tissue samples, with a coefficient of variation (CV) typically less than 10%.
Analyte Details: Comprehensive information on each detected sugar, including name, abbreviation, chemical formula, molecular weight, and CAS number.
Determination of monosaccharides in Lycium barbarum fruit polysaccharide by an efficient UHPLC-QTRAP-MS/MS method
Journal: Phytochemical Analysis
Published: 2021
Background
Herbal plants have played a crucial role in human life and traditional medicinal systems for centuries. Many of these plants possess medicinal potential and can be consumed as food. Notably, the genus Eremostachys Bunge was used for treating poisoning and its fresh leaves were consumed as vegetables. The genus Ferula was utilized for making gums and has exhibited antiseptic, anticancer, and anti-epilepsy properties. Additionally, species from the genus Perovskia were used as tonics in Iranian folk medicine. Lycium barbarum, widely used in Chinese folk medicine, is known for its therapeutic effects on blood sugar and lipid regulation, immune response enhancement, anti-tumor and antioxidant properties, and protection of the liver and kidneys. L. barbarum fruit contains various chemical components, with polysaccharides being the main active ingredient. Despite its significance, detailed reports on the chemical analysis and comparison of L. barbarum polysaccharides (LBP) from different locations are limited. Therefore, analyzing LBP composition is essential for understanding its biological mechanisms.
Technical Methods
Chemicals and Reagents
L. barbarum samples were collected from various locations, dried, and identified botanically. Various monosaccharides and reagents were used for the study, including galactose, arabinose, mannose, rhamnose, xylose, ribose, glucose, ammonium acetate, and PMP for pre-column derivatization. Chromatography-grade solvents and double distilled water were also utilized.
Standard Solution Preparation
A mixed monosaccharide standard solution was prepared and diluted to required concentrations for calibration. The solution was filtered and stored at 4°C.
Sample Solution Preparation
- LBP Extraction:
- L. barbarum powder was defatted with petroleum ether and extracted with methanol.
- The residue was extracted with boiling water, filtered, and the extract was condensed and precipitated with methanol.
- The polysaccharide precipitate was obtained by centrifugation and dried to yield LBP.
- LBP Hydrolysis:
- LBP powder was hydrolyzed with hydrochloric acid at 110°C for 1 hour.
- The solution was neutralized and prepared for analysis.
- PMP Derivatization:
- LBP solution was reacted with PMP and sodium hydroxide.
- The solution was extracted with chloroform and filtered before UHPLC-QTRAP-MS/MS analysis.
UHPLC-QTRAP-MS/MS Instrumentation and Conditions
The UHPLC-QTRAP-MS/MS system was utilized for glycomics analysis, offering high sensitivity and specificity in detecting glycan structures and monosaccharide derivatives. Chromatographic separation using specific columns and optimized mobile phases facilitated detailed analysis of glycan profiles and metabolomics. Chromatographic separation was performed using a specific column with a mobile phase of ammonium acetate and acetonitrile. The positive ion MRM mode was employed for detecting the target compounds, with optimized parameters for MS analysis.
Method Validation
The method was validated for linearity, detection limit, precision, stability, repeatability, and recovery. Calibration curves were established, and the LOD and LOQ were determined. Precision was assessed by RSD, and repeatability was evaluated by multiple determinations of hydrolyzed monosaccharides. Stability was tested by analyzing samples at different times, and recovery was validated by adding standard solutions to samples.
Multivariate Statistical Analysis
Principal component analysis (PCA) and partial least square discriminant analysis (PLS-DA) were used to evaluate variations among monosaccharides in L. barbarum samples. Technique for order of preference by similarity to ideal solution (TOPSIS) was employed to rank the samples based on their quality.
Results
Quantitative analysis revealed significant differences in monosaccharide content among L. barbarum samples from different locations, with Ningxia samples having the highest content. Glucose was the most abundant monosaccharide, followed by galactose and mannose. Multivariate analysis indicated that monosaccharide content could distinguish the origin of the samples, with Ningxia samples showing better quality. The study established a novel, rapid, and sensitive UHPLC-QTRAP-MS/MS method for detecting monosaccharides in L. barbarum, with hydrochloric acid proving more efficient for hydrolysis than TFA. This method can be used to evaluate the quality of L. barbarum and other medicinal plants containing polysaccharides, although further detection methods are needed for other monosaccharides.
 Multiple reaction monitoring (MRM) chromatograms of seven monosaccharides
Multiple reaction monitoring (MRM) chromatograms of seven monosaccharides
 Multivariate statistical analyses of samples. (a) Principal component analysis (PCA) score chart. (b) Partial least square discriminant analysis (PLS-DA) score chart. (c) Variable importance projection (VIP)
Multivariate statistical analyses of samples. (a) Principal component analysis (PCA) score chart. (b) Partial least square discriminant analysis (PLS-DA) score chart. (c) Variable importance projection (VIP)
Reference
- Xu, Jun, et al. "Determination of monosaccharides in Lycium barbarum fruit polysaccharide by an efficient UHPLC‐QTRAP‐MS/MS method." Phytochemical Analysis 32.5 (2021): 785-793.






