Plant Primary Metabolites Analysis Service
At Creative Proteomics, we provide a comprehensive LC-MS/MS-based platform for the accurate profiling of primary metabolites in plants. Tailored for both academic and industrial research applications, our service enables detailed analysis of core metabolic pathways, including carbohydrates, amino acids, organic acids, and nucleotides.
Our Plant Metabolomics Platform Features:
- Technology: High-sensitivity Liquid Chromatography–Tandem Mass Spectrometry (LC-MS/MS)
- Method: Optimized extraction protocols for plant tissues, cells, and fluids
- Flexibility: Targeted and untargeted workflows adaptable to diverse species and experimental goals
- Coverage: Broad detection of key metabolite classes involved in growth, development, and stress responses
Whether you're decoding plant-microbe interactions or screening for metabolic biomarkers, our service delivers reproducible, high-resolution data to accelerate your discoveries with confidence.
Submit Your Request Now
×- Background
- Service Overview
- Metabolite Coverage
- Platform
- Workflow
- Sample Requirement
- Delivery & Demo
- FAQ
- Case Study
- Publication
What Are Plant Primary Metabolites?
Plant primary metabolites are fundamental organic compounds required for plant growth, development, and survival. They directly participate in essential biological processes such as energy generation, structural maintenance, and metabolic regulation.
These metabolites fall into several main categories:
- Carbohydrates like glucose and sucrose serve as primary energy sources and structural materials.
- Amino acids are the building blocks of proteins that drive cellular repair, enzyme activity, and biochemical signaling.
- Organic acids, including citric and malic acid, play key roles in cellular respiration and energy cycles like the Krebs cycle.
- Fatty acids contribute to membrane integrity and act as long-term energy reserves, supporting plant stress tolerance and adaptability.
- Studying primary metabolites is vital for understanding plant physiology, enhancing agricultural productivity, and improving crop quality. These molecules also influence food nutrition and how plants interact with their environment.
At Creative Proteomics, we offer specialized services for analyzing plant primary metabolites, helping researchers and industry teams generate meaningful, data-driven insights.
Plant Primary Metabolites Analysis Offered by Creative Proteomics
We provide comprehensive services that cover the full spectrum of primary metabolite analysis:
Qualitative and Quantitative Analysis
Measure the presence and concentration of individual metabolites in plant tissues.
Metabolomic Profiling
Profile multiple metabolite classes simultaneously to capture the overall metabolic status of the plant.
Metabolite Identification
Use advanced detection technologies to identify unknown compounds within complex plant matrices.
Customized Analysis Solutions
Tailor workflows and reporting formats to meet your specific research or industrial project goals.
List of Detected Plant Primary Metabolites
We support detection of a wide range of primary metabolite classes and representative compounds:
| Metabolite Class | Example Metabolites |
|---|---|
| Carbohydrates | Glucose, Fructose, Sucrose, Galactose, Ribose, Xylose, Mannose |
| Amino Acids | Glutamic Acid, Alanine, Glycine, Valine, Leucine, Isoleucine, Serine, Aspartic Acid, Phenylalanine, Tryptophan, Threonine, Methionine |
| Organic Acids | Citric Acid, Malic Acid, Lactic Acid, Acetic Acid, Oxalic Acid, Fumaric Acid, Succinic Acid, Ascorbic Acid |
| Fatty Acids | Palmitic Acid, Linoleic Acid, Oleic Acid, Stearic Acid, Alpha-Linolenic Acid |
| Nucleotides | Adenosine Triphosphate (ATP), Guanosine Triphosphate (GTP) |
Metabolomics Services
Creative Proteomics provides both untargeted and targeted metabolomics services designed to reveal biological mechanisms and metabolic pathways:
Untargeted Metabolomics identifies differentially expressed metabolites using broad screening followed by standard verification.
Targeted Metabolomics validates key findings through precise quantification, supporting applications such as biomarker discovery and mechanism-of-action studies.
Download our brochure to explore all available solutions.
Technology Platforms for Plant Primary Metabolites Assay
We utilize a range of validated analytical technologies to support diverse plant research goals:

Gas Chromatography–Mass Spectrometry (GC-MS)
Best for volatile compounds. GC-MS pairs a gas chromatograph with a mass spectrometer, offering high detection sensitivity and throughput. Critical parameters include carrier gas flow, column type, and ionization mode.

Liquid Chromatography–Mass Spectrometry (LC-MS)
Ideal for non-volatile and thermally sensitive metabolites. LC-MS delivers excellent reproducibility, broad compound coverage, and high quantitation accuracy. Ionization methods such as ESI and APCI are selectable based on compound polarity.

High-Performance Liquid Chromatography (HPLC)
A robust platform for separation and quantification. With options for UV/Vis or PDA detection, HPLC is cost-effective and flexible for various plant matrices.
Workflow

Sample Requirements for Plant Primary Metabolites Analysis
Please follow the guidelines below for optimal sample preparation:
| Sample Type | Recommended Volume | Preservation Method | Storage Conditions |
|---|---|---|---|
| Fresh Plant Tissue | 5–10 g | Snap freeze in liquid nitrogen | -80°C |
| Dried Plant Material | 1-2 g | Store in a desiccator | Room temperature |
| Aqueous Extracts | 1–2 mL | Refrigerate immediately | 4°C |
| Plant Juices | 1–2 mL | Refrigerate | 4°C |
| Root Samples | 5–10 g | Rinse with distilled water, freeze | -80°C |
| Leaf Samples | 5–10 g | Snap freeze or dry quickly | -80°C or room temperature |
| Fruit Samples | 5–10 g | Refrigerate or freeze immediately | 4°C or -80°C |
| Seeds | 5 g | Dry in a desiccator | Room temperature |
| Plant Cell Cultures | 1-2 mL | Maintain in growth medium | 4°C or room temperature |
| Tissue Culture Samples | 1-2 mL | Store in growth medium | 4°C |
Important Notes:
- Prepare and freeze samples quickly to prevent degradation.
- Label all samples clearly with collection details and any treatments applied.
- Contact us for advice if your sample type is not listed.
What You'll Receive in the Final Report
We deliver a comprehensive, easy-to-interpret report in both Excel and PDF formats. It includes:
- Raw data and annotated spectra
- Instrument settings and parameters
- Full calculation workflows and quality metrics
Demo
Frequently Asked Questions
How are primary metabolites extracted from plant samples?
Extraction methods depend on the type of metabolite being analyzed. Common approaches include:
- Solvent extraction using methanol-water mixtures
- Liquid-liquid extraction for partitioning metabolites based on polarity
- Solid-phase extraction to isolate specific compound classes
These processes are optimized for yield and purity and typically involve homogenization, centrifugation, and filtration steps to prepare clean extracts for analysis.
Why is metabolomic profiling important in plant research?
Metabolomic profiling offers a full snapshot of a plant's metabolic state. It helps researchers:
- Detect biochemical changes linked to stress, development, or treatment
- Understand how pathways shift under different conditions
- Identify metabolic markers for breeding or engineering purposes
This systems-level view makes it easier to interpret complex biological interactions and optimize traits like growth rate, yield, or stress resistance.
What role do primary metabolites play in stress responses?
Primary metabolites are key players in how plants adapt to stress. For example:
- Amino acids support the production of protective proteins
- Sugars supply the energy needed for repair and recovery
Tracking these metabolites reveals how plants respond to drought, salinity, pathogens, and other challenges—offering insights into resilience mechanisms and targets for crop improvement.
Can your analysis distinguish between metabolite isomers or isoforms?
Yes. Our LC-MS and NMR platforms provide high-resolution analysis that can distinguish between isomers—compounds with the same molecular weight but different structures.
- LC-MS separates compounds based on differences in mass-to-charge ratios (m/z).
- Using ionization methods like ESI or APCI, we enhance both sensitivity and selectivity.
- NMR spectroscopy adds structural detail, identifying the position of functional groups and atomic connectivity.
What types of plant samples are accepted for metabolite analysis?
We accept a wide variety of sample types, including:
- Fresh or dried plant tissues (leaves, roots, fruits, seeds)
- Plant cell cultures or tissue cultures
- Plant extracts, juices, or culture media
If you're unsure whether your sample type is compatible, our technical team will evaluate and recommend the best processing method to ensure accurate results.
How much sample is needed for analysis?
The amount required depends on the sample matrix, but general guidelines include:
- Fresh/Dried Tissue: 5–10 grams
- Extracts or Juices: 1–2 mL
- Cell Cultures: 1–2 mL
All samples should be labeled clearly and preserved according to our sample preparation guide. For unique matrices, we offer one-on-one support during submission.
What do I receive in the final report?
Our comprehensive data report includes:
- Quantified metabolite levels in µM or µg/mg
- Raw data files, including chromatograms and spectra
- Statistical plots (e.g., PCA, volcano plots, heatmaps)
- Pathway enrichment analysis with biological interpretation
Reports are delivered in Excel and PDF format for easy review and integration into your workflow or publication.
Can the service be customized for my specific research needs?
Yes. We offer flexible study designs to support:
- Specific metabolite panels (e.g., carbohydrate-rich profiling)
- Species-specific optimization (e.g., Arabidopsis, maize, tea, rice)
- Time-course studies or stress condition comparisons
Simply share your project goals and our team will tailor the analysis plan accordingly.
How do you ensure data accuracy and reproducibility?
All analyses are performed under strict quality control using:
- Internal standards and technical replicates
- Validated instrument calibration protocols
- QC samples for batch-to-batch consistency
We maintain CVs under 10% for most metabolites, ensuring reliable, publishable data.
Can this service help me identify biomarkers or pathway targets?
Absolutely. Many clients use our platform for:
- Biomarker discovery (e.g., drought resistance indicators)
- Functional metabolite mapping
- Screening metabolic shifts in genetically modified plants
We also offer pathway annotation tools to highlight functional clusters and regulatory nodes in your dataset.
Do you offer bioinformatics or statistical consulting?
Yes. We provide optional downstream analysis services, including:
- Multivariate statistics (PCA, PLS-DA)
- Differential metabolite screening
- Correlation analysis with phenotype or gene expression
Our bioinformatics experts can help interpret your results and guide next steps in experimental design.
Can I combine primary metabolite analysis with other omics data?
Yes. Many clients integrate our primary metabolite data with:
- Transcriptomics (RNA-Seq)
- Proteomics
- Secondary metabolite profiling
We support multi-omics project design and can align your datasets for joint pathway or network analysis.
Learn about other Q&A.
Customer Case Study: Revealing Plant Growth-Promoting Metabolites in Microalgae

https://doi.org/10.3390/plants12030651
- Background
- How Creative Proteomics Helped
- Results & Impact
- Why It Matters
To explore the untapped potential of rhizosphere-associated microalgae in plant growth promotion, researchers at iBET and the University of Waterloo investigated a novel strain of Micractinium rhizosphaerae isolated from Portuguese soil. Their goal was to identify phytohormones and other bioactive metabolites that could explain the strain's beneficial effects on tomato plants.
The research team needed a sensitive, untargeted metabolomics platform to analyze complex exudates from Micractinium rhizosphaerae. Creative Proteomics delivered high-resolution, LC-MS-based profiling of the microalgal supernatant, including:
- Sample preparation from culture broth
- Untargeted LC-MS in both positive and negative ionization modes
- Peak annotation, metabolite identification, and quantitative data output
- Identification of key phytohormones such as indole-3-acetic acid (IAA), salicylic acid (SA), jasmonates, and abscisic acid (ABA)
The analysis detected over 9000 ion peaks and successfully identified dozens of metabolites, including several plant hormones previously unreported in this species. This allowed the researchers to:
- Confirm the presence of IAA, SA, JA, and ABA in the algal exudates
- Correlate hormone profiles with tomato root colonization and biomass increase
- Support their proposal of M. rhizosphaerae as a novel plant-growth-promoting algae (PGPA) species
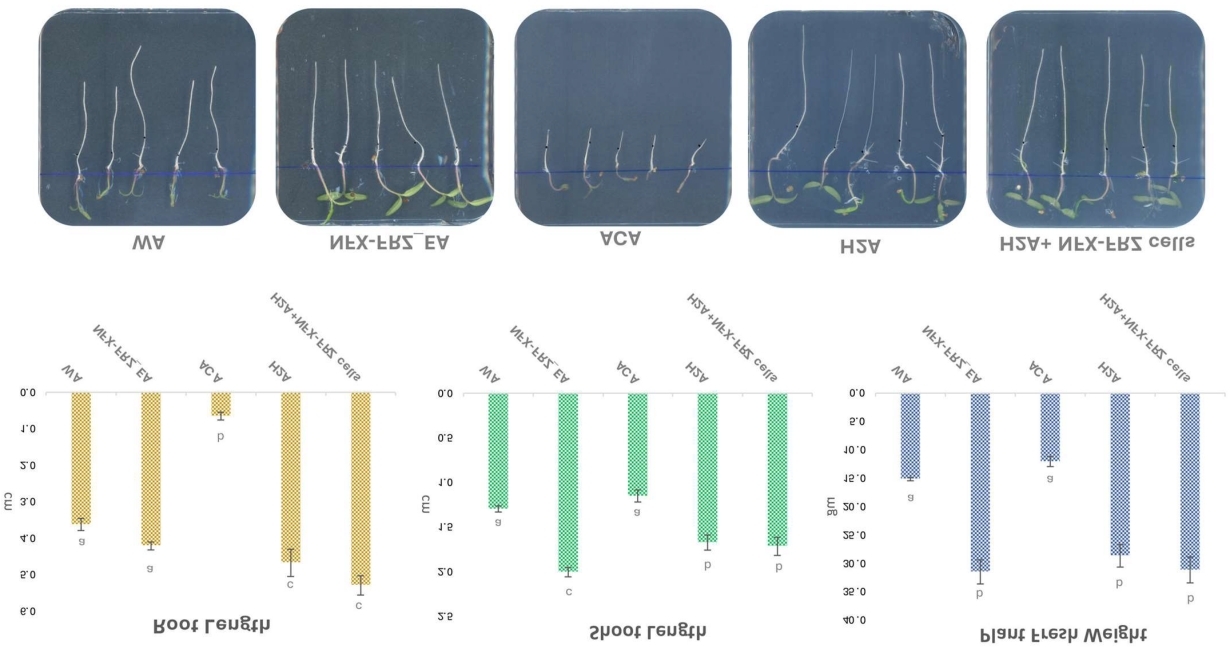 Tomato plant growth under different treatments (including microalgal exudates)
Tomato plant growth under different treatments (including microalgal exudates)
This metabolic insight was critical in validating the biofertilizer potential of the new species and laid the foundation for future plant–microalgae interaction studies.
By providing robust, publication-quality metabolomic data, Creative Proteomics helped accelerate the discovery and validation of novel plant-growth-promoting mechanisms. The results supported the publication of a high-impact study in Plants
Our Plant Primary Metabolites Analysis Publication

- Physiological, transcriptomic and metabolomic insights of three extremophyte woody species living in the multi-stress environment of the Atacama Desert. Gajardo, Humberto A., et al. Journal: Planta. 2024. https://doi.org/10.3390/antiox9111098
- Combined omics approaches reveal distinct mechanisms of resistance and/or susceptibility in sugar beet double haploid genotypes at early stages of beet curly top virus infection. Galewski, Paul J., et al. Journal: International Journal of Molecular Sciences. 2023. https://doi.org/10.3390/ijms241915013
- Plant Growth Promotion, Phytohormone Production and Genomics of the Rhizosphere-Associated Microalga, Micractinium rhizosphaerae sp. nov. Quintas-Nunes, Francisco, et al. Journal: Plants. 2023. https://doi.org/10.3390/plants12030651
- Summative and ultimate analysis of live leaves from southern US forest plants for use in fire modeling. Matt, Frederick J., Mark A. Dietenberger, and David R. Weise. Journal: Energy & Fuels 2020. https://dx.doi.org/10.1021/acs.energyfuels.9b04495
- Detailed analysis of agro-industrial byproducts/wastes to enable efficient sorting for various agro-industrial applications. Priyanka, Govindegowda, et al. Journal: Bioresources and Bioprocessing. 2024. https://doi.org/10.1186/s40643-024-00763-7