Cysteine Profiling Service using LC-MS/MS
Unlock the full picture of cysteine biology with Creative Proteomics' next-generation cysteine modification proteomics platform. Leveraging extensive project experience, our R&D team has expanded beyond classic S-nitrosylation to deliver a comprehensive cysteine post-translational modification analysis service that captures:
- S-palmitoylation and other thio-acylations
- S-nitrosylation and global reversible oxidation
- S-glutathionylation, S-sulfenylation (-SOH), S-sulfination (-SO₂H)
- Free-thiol (unmodified) proteome mapping
Submit Your Request Now
×
- Raw LC-MS + QC metrics – full transparency, straight from our Orbitrap/timsTOF platforms
- Curated PTM site tables – high-confidence localisation, fold-changes, statistics
- Publication-ready visuals – spectra, volcano plots, pathway maps
- Background
- Service Overview
- Methods&Workflow
- Advantage
- Sample Requirement
- Delivery&Demo
- FAQ
- Case Study
- Publication
Introduction: Why Cysteine PTMs Matter
Cysteine stands out among proteinogenic amino acids because its thiol (-SH) side chain is exquisitely redox-sensitive, acting as both a catalytic nucleophile and a molecular switch that responds to oxidative cues. A single cysteine post-translational modification (PTM)—be it reversible S-nitrosylation or the formation of an intramolecular disulfide—can realign a protein's conformation, alter its interaction landscape, and ultimately reprogram entire signaling cascades. In drug-discovery settings, these redox-driven switches modulate target engagement, off-target liabilities, and even compound stability, making cysteine PTMs a powerful—but often under-measured—layer of functional regulation.
Despite their importance, cysteine modifications are notoriously transient and present at low stoichiometry, meaning traditional proteomics workflows miss a substantial fraction of the actionable data. Purpose-built cysteine post-translational modification profiling therefore provides a sharper lens for researchers investigating stress responses, kinase regulation, or covalent inhibitor mechanisms. By capturing the dynamic redox landscape instead of static protein abundance alone, scientists gain a more predictive understanding of cellular behavior—insights that are increasingly pivotal for academic discovery and pre-clinical R&D alike.
Common Cysteine Modifications at a Glance
Cysteine's thiol is chemically versatile, giving rise to a spectrum of post-translational modifications (PTMs) that act as rapid "switches" in redox and signaling networks. Below are five of the most biologically prominent—each detectable through our cysteine profiling workflow.
| Modification | Chemical Feature | Typical Functional Outcome | Representative Evidence |
|---|---|---|---|
| S-Nitrosylation (SNO) | Addition of an –NO group to the thiol, forming S-nitrosothiol | Modulates enzyme activity, protein–protein interactions, and redox cycling; often reversible via denitrosylases | TRAP1 chaperone S-nitrosylation reshapes conformational dynamics and redox communication (Papaleo et al., 2023. DOI: https://doi.org/10.1038/s41419-023-05780-6) |
| S-Palmitoylation (S-Acylation) | Thioester linkage of a C16 fatty acid (or related acyl chain) | Anchors proteins to membranes, controls trafficking and half-life; dynamic via DHHC transferases and thioesterases | Heart proteome study mapped >1,000 palmitoylated sites, underscoring prevalence of this lipid PTM (Miles et al., 2021. DOI: https://doi.org/10.1016/j.yjmcc.2021.02.007) |
| Disulfide Bond Formation | Intra- or intermolecular –S–S– linkage between two cysteines | Stabilizes tertiary/quaternary structure, acts as redox‐sensing "lock" that can be broken under reducing conditions | Classic in ER folding and redox signaling; readily captured in non-reducing gel or MS workflows |
| S-Sulfenylation (-SOH) | Oxidation to sulfenic acid, a fleeting intermediate | Serves as a redox "gateway" that can resolve to disulfide, sulfinic/sulfonic acid, or glutathionylation; regulates kinase and phosphatase activity | Global chemoproteomic mapping revealed >1,000 sulfenylated sites in human cells (Yang et al., 2014. DOI: https://doi.org/10.1038/ncomms5776) |
| S-Glutathionylation (SSG) | Mixed disulfide between protein cysteine and glutathione | Shields catalytic thiols from irreversible oxidation and tunes signaling cascades (e.g., NF-κB) | Isotopically labeled "click-GSH" workflow quantified proteome-wide SSG changes (VanHecke et al., 2020. DOI: https://doi.org/10.1002/cbic.201900528) |
Why it matters for your project: Each PTM leaves a unique chemical "fingerprint" that our selective enrichment chemistries and high-resolution LC-MS/MS can capture. Whether you need S-nitrosylation analysis for oxidative stress models or palmitoylation mapping
Research & Drug-Discovery Challenges
High-quality cysteine PTM data are difficult to obtain because the chemistry we're trying to capture is both fleeting and sparse. In whole-cell lysates,<5 % of any given cysteine population may carry a specific modification at steady state; sample handling further skews this low stoichiometry by introducing thiol oxidation artifacts or causing labile PTMs (e.g., S-nitrosylation) to collapse before analysis. Even when the modification survives, a single reactive probe rarely labels every accessible cysteine: head-to-head comparisons show pronounced probe- and warhead-specific biases that leave swaths of the "cysteinome" invisible to any one workflow.
Key Pain Points for Researchers
Low stoichiometry & dynamic range – Modified cysteines are often present at sub-percent levels, demanding highly efficient enrichment and ultra-sensitive LC-MS/MS.
Probe coverage & selectivity – Comparative datasets (e.g., CysDB) reveal that different iodoacetamide- or dimedone-based chemistries capture overlapping but non-identical site sets, complicating global profiling studies.
Sample-induced artifacts – Air exposure, residual metal ions, or non-ideal buffers can oxidize free thiols, masking true in-vivo PTM states if not rigorously controlled.
MS/MS fragmentation pitfalls – Click-based capture tags can fragment unpredictably, reducing peptide ID rates unless acquisition methods and search algorithms are optimized (Yan et al., 2022). We minimise this issue by adopting short-linker biotin-DBCO tags and optimised library-search parameters.
Bioinformatic localization – Distinguishing closely spaced cysteines or multiple oxidative states on the same residue requires specialized scoring models and manual validation checkpoints.
Because of these hurdles, many labs see high false-negative rates or irreproducible hit lists when attempting in-house assays. A dedicated Cysteine Profiling Service—equipped with modular probe panels, inert-atmosphere sample prep, and purpose-built informatics—solves these challenges and delivers reliable, quantifiable insights that directly inform covalent inhibitor design, stress-response studies, and redox signaling projects.
Service Overview — Creative Proteomics Cysteine Profiling Platform
Creative Proteomics has built a Cysteine Profiling Service specifically for redox-sensitive residues, combining chemistry-matched enrichment probes with state-of-the-art LC-MS/MS cysteine analysis and bespoke bioinformatics. The platform is modular, letting clients interrogate a single PTM (e.g., S-nitrosylation) or run a broad "cysteinome" screen in one streamlined project.
| Core Component | What We Do | Why It Matters for Your Study |
|---|---|---|
| Selective Probe Panel | Iodoacetamide-alkyne, dimedone, maleimide, and acyl-switch chemistries applied under inert atmosphere to preserve labile PTMs. | Captures complementary cysteine subsets and minimizes false negatives (Backus et al., 2016. DOI: 10.1038/nature18002). |
| Precision LC-MS/MS | Orbitrap Exploris 480 and timsTOF HT operated at sub-ppm mass accuracy with data-independent acquisition (DIA) or TMT-based multiplexing. | Achieves >25,000 unique cysteine site IDs per experiment (extended gradients can exceed 30,000). |
| Automated Data Pipeline | PTM-Site Localizer™ software scores modification sites, filters by localization probability, and maps hits onto KEGG/Reactome pathways. | Fast turn-around of actionable tables, spectra, and pathway visuals—ready for grant figures or medicinal-chemistry follow-up. |
| Project Management & Consulting | Dedicated Ph.D. scientist aligns probe choice, MS method, and statistics with your biological question; optional integration with global PTM or proteome workflows. | Ensures each dataset dovetails with broader mechanistic studies, saving you iterative rounds of optimization. |
When your project requires multi-layered PTM insight, the cysteine platform plugs directly into our broader [Protein Post-Translational Modification Analysis Service] for phospho-, acetyl-, or ubiquitin-level context—no extra sample prep needed.
Technology & Workflow — Step-by-Step
Below is the standardized cysteine enrichment workflow we apply to every project. Each stage is performed under rigorously controlled, oxygen-limited conditions to preserve labile thiol chemistry and maximize PTM site mapping depth.
| Step | Key Actions | Technical Highlights |
|---|---|---|
| 1. Sample Receipt & Pre-Processing | Verify protein concentration, buffer composition, and reducing-agent history; rapidly snap-freeze aliquots under nitrogen. | Accepts cells, tissues, or purified protein (≥100 µg total protein recommended). |
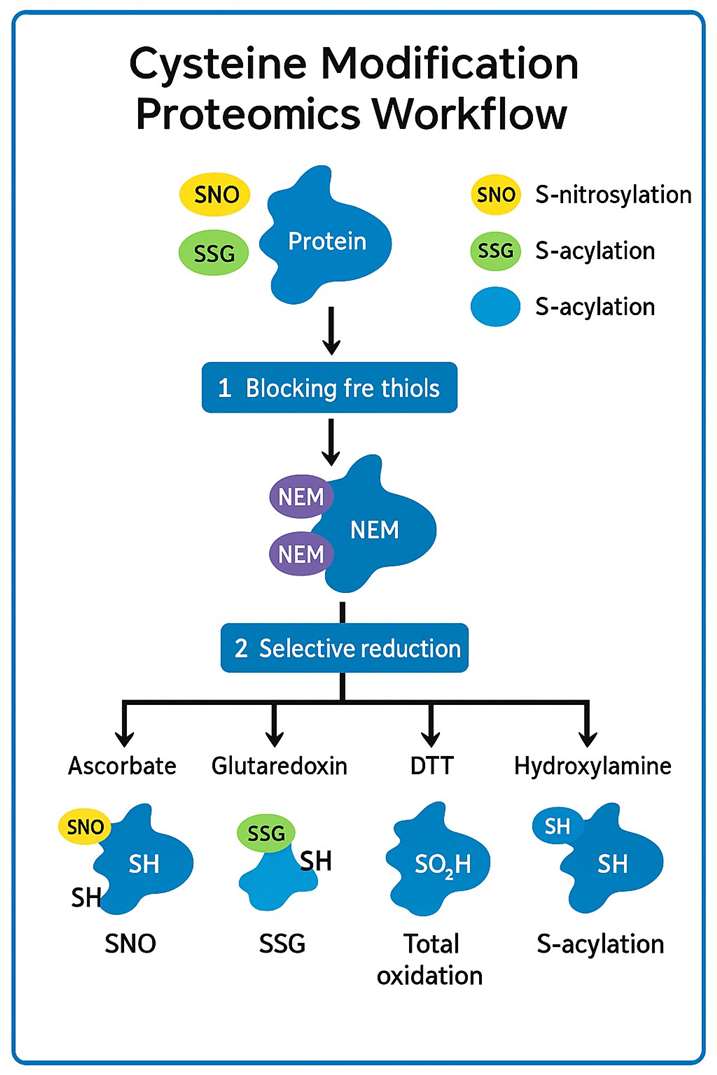
| 2. Thiol Blocking ("Freezing" Native Redox State) | Alkylate unmodified –SH groups with light/dark iodoacetamide (IAA) or N-ethylmaleimide to prevent ex-vivo oxidation. | Dual-label option enables relative quantification of pre-existing vs newly formed PTMs. |
| 3. Selective PTM Exchange / Probe Tagging | Apply chemistry-matched probes (e.g., ascorbate-based "biotin-switch" for S-nitrosylation, acyl-switch for palmitoylation, dimedone-alkyne for sulfenylation). | Modular panel captures multiple cysteine PTMs in parallel. |
| 4. Affinity Enrichment | Perform CuAAC "click" to biotin, then pull-down on streptavidin or NeutrAvidin beads; extensive washes remove non-specific binders. | ≥200-fold enrichment over unenriched lysate. |
| 5. Proteolytic Digestion & Peptide Clean-Up | On-bead Lys-C/trypsin digest, desalting via SDB-RPS StageTips. | Generates MS-compatible peptides with conserved probe adducts. |
| 6. High-Resolution LC-MS/MS | Nano-LC separation (EvoSep or EASY-nLC1200) → Orbitrap MS (Exploris 480) or timsTOF HT with data-independent acquisition (g-DIA) or TMT-multiplexed SPS-MS³. | Sub-ppm mass accuracy; ≤1 h gradients for high throughput. |
| 7. Bioinformatic PTM Site Mapping | Raw→mzML conversion → PTM-aware search (ionQuant, PTM-Site Localizer™) → localization probability scoring → Stoichiometry & fold-change calculations. | Spectral-library or library-free DIA workflows supported. |
| 8. Report Generation & Expert Review | Deliverables: Excel hit list, annotated spectra (PDF), volcano plots, KEGG/Reactome pathway maps, and methodological appendix for Methods sections. | Interactive dashboard (optional) for drill-down exploration. |
By integrating stringent thiol preservation, chemistry-selective capture, and sub-ppm Orbitrap MS acquisition, our pipeline consistently identifies >25 k unique cysteine sites with high localization confidence—transforming the traditionally elusive cysteine redox landscape into actionable, quantitative datasets.
Advantages of Partnering with Creative Proteomics
1. Unrivalled Cysteine-Modification Coverage
Profile seven major reversible chemistries in one project: S-palmitoylation and other thio-acylations, S-nitrosylation, S-glutathionylation, S-sulfenylation (-SOH), S-sulfination (-SO₂H), persulfidation (-SSH), global reversible oxidation, plus free-thiol mapping.
2. Dual Labelling Strategies for Depth and Flexibility
Biotin-exchange workflow (streptavidin enrichment) — no cap on sample number, very low reagent cost, and stronger affinity than antibody-based pull-downs, giving the deepest site recovery for discovery screens.
TMT multiplexing — up to 16 conditions in one LC-MS injection for accurate relative quantification when sample numbers are defined.
Our team selects—or combines—these chemistries to match study size, quantitative needs and budget.
3. Ultra-High Sensitivity, Validated by Site-Level FDR
Orbitrap Exploris 480 and timsTOF HT routinely quantify > 25 000 unique cysteine sites at ≤ 1 % site-level FDR, capturing low-stoichiometry events such as covalent-drug engagement or stress-induced redox switches.
4. Control-Driven Experimental Design
Each biological sample is paired with an internal control to eliminate system bias. Strict localisation scoring and a global 1 % false-discovery filter ensure every reported site is robust enough for publication review.
5. Rapid Turnaround with Expert Guidance
EvoSep high-throughput LC, automated data pipelines and a dedicated Ph.D. project manager keep timelines short and communication seamless—from probe choice to figure-ready deliverables.
6. One-Stop PTM Expansion and Scalable Pricing
The cysteine workflow dovetails directly with our phospho-, acetyl- and ubiquitin-omics services, enabling facile systems-biology add-ons. Pricing tiers cover single samples, TMT multiplexes or 96-well plate screens, so you can start lean and scale fast.
Sample Requirements & Submission Guidelines
| Sample Category | Sub-type Examples | Minimum Amount | Notes |
|---|---|---|---|
| Solid Samples | Common animal tissues (liver, muscle, etc.) | ≥ 300 mg | Flash-freeze in liquid N₂; ship on dry ice. |
| Soft plant tissues (leaf, flower, fruit) | ≥ 300 mg | Remove excess moisture; snap-freeze. | |
| Bacterial biomass | ≥ 500 mg wet pellet | Pellet at 4 °C, discard supernatant, freeze immediately. | |
| Fungal mycelium / spores | ≥ 8 g wet weight | Rinse quickly with PBS, blot dry, snap-freeze. | |
| Cultured cells | ≥ 8 × 10⁷ cells | Wash twice with PBS, pellet, flash-freeze. | |
| Liquid Samples | Serum / plasma | ≥ 1 mL | Use anticoagulant-free (serum) or EDTA (plasma); avoid haemolysis. |
| Milk / plant sap | ≥ 5 mL | Clarify by low-speed spin to remove debris. | |
| Urine | ≥ 50 mL | Collect mid-stream; add protease inhibitors if delay > 2 h. | |
| Other biofluids (CSF, synovial fluid, etc.) | ≥ 500 µL | Filter large particulates; keep on ice before freezing. | |
| Secreted-protein media (cell-culture or tissue explant) | ≥ 10 mL | Centrifuge 300 × g, 10 min; collect supernatant only. |
Buffer compatibility
Avoid glycerol > 5 %, strong detergents (e.g., SDS > 0.1 %), and high salt > 250 mM.
Do not add reducing agents (DTT, TCEP) or alkylators (IAA, NEM); our workflow introduces controlled labelling.
Packaging & Shipping
- Aliquot samples to minimise freeze-thaw cycles (≤ 500 µL or ≤ 500 mg per tube).
- Label tube tops and sides with sample ID, matrix, and weight/volume.
- Freeze at −80 °C and ship overnight on dry ice (≥ 5 kg for international parcels).
- Include the completed Cysteine-PTM Sample Submission Form inside the package.
Turnaround Options remain unchanged (Standard / Priority / High-throughput); the project clock starts once samples pass QC for weight, protein concentration and buffer compatibility.
Deliverables & Data Interpretation Support
| Deliverable | Format | What It Contains | How It Helps You |
|---|---|---|---|
| Raw MS files | .raw (Thermo) or .d (Bruker) | All centroid and profile spectra from Orbitrap / timsTOF acquisition | Full transparency—re-process with your preferred pipeline or archive for journal reviewers |
| Peptide & Site Lists | Excel + .csv |
| Quickly filter by fold-change, confidence, or pathway to pinpoint hits relevant to your hypothesis |
| Annotated MS/MS Spectra | PDF bundle | Fragment-ion maps with b-/y-series assignments and probe-specific reporter ions | Facilitates peer-review and supports figure preparation |
| Quantitative Summary Tables | Excel pivot tables | Normalized abundance, statistical tests (ANOVA/limma), volcano-plot source data | Identifies statistically significant PTM shifts across treatments or time points |
| Pathway & Network Maps | Interactive HTML or static PNG | Overlay of modified proteins on KEGG / Reactome pathways; Cytoscape network of PTM hotspots | Visualizes how cysteine modifications reshape signaling or metabolic circuits |
| Methodological Appendix | Word / PDF | Detailed protocols, instrument settings, search-engine parameters, and QC metrics | Paste directly into Materials & Methods sections of manuscripts or grant apps |
| Optional Dashboard | Secure web portal | Drill-down view of spectra, plots, and site metadata; export functions | Accelerates data exploration for multi-disciplinary teams |
Site-level validation advice (e.g., targeted PRM assays, mutagenesis controls).
Integration support with our global PTM or whole-proteome datasets when broader context is required.
DEMO Results Snapshot
Frequently Asked Questions (FAQs)
What is the minimum amount of material I should send?
For reliable site coverage, please provide at least 300 mg of solid tissue (animal or plant), 500 mg of bacterial biomass, 8 g of fungal material, 8 × 10⁷ cultured cells, or the liquid-sample volumes listed above (e.g., 1 mL serum, 50 mL urine). Smaller inputs can be processed, but depth and quantitation may decline—contact us to discuss pilot options.
Which cysteine modifications can you monitor in a single experiment?
Our probe panel captures all major reversible chemistries—S-nitrosylation, S-palmitoylation (and other thio-acylations), S-glutathionylation, S-sulfenylation, S-sulfination, persulfidation, and global reversible oxidation. You can profile one PTM class or combine several in the same run.
How sensitive is the workflow?
With selective enrichment and high-resolution Orbitrap/timsTOF MS, we routinely detect low-stoichiometry sites that represent well under 1 % of the total cysteine pool—provided recommended input amounts and clean buffers are used.
Can I run multiple samples together for relative quantification?
Absolutely. We support TMT multiplexing for up to 16 conditions as well as data-independent acquisition (DIA) for larger cohorts when broad coverage is the priority.
What formats will my results arrive in?
You receive raw MS files, Excel/CSV hit lists with localization probabilities, annotated spectra (PDF), pathway visualisations, and a detailed methods appendix that can be dropped straight into manuscripts or grant applications.
How secure are my data and intellectual property?
All files are transferred through an encrypted client portal with role-based access, and we are happy to sign NDAs. You retain full ownership of every dataset; we never share client information without written permission.
Can the data be integrated with other omics layers?
Yes. Our informatics pipeline can merge cysteine PTM ratios with phosphoproteomics, acetylomics, or whole-proteome data—whether generated by our lab or yours—to give a comprehensive systems-level view.
Do you accommodate large screening projects?
We do. The platform scales from single proof-of-concept samples to 96-well plate screens. Turnaround and pricing are scoped case-by-case to match your study's size and complexity.
Are the results suitable for publication?
Definitely. Every report includes complete raw spectra, QC metrics, and methodological detail that meet journal peer-review standards for transparency and reproducibility.
Learn about other Q&A about other technologies.
Case Study Spotlight — From Cysteine Oxidation Maps to Drug-Target Insight

- Overview
- Results
- Conclusion
- Why it matters
A recent investigation in Journal of Translational Medicine used high-coverage cysteine redox proteomics to interrogate pancreatic-cancer biology (Tan et al., 2024. DOI: https://doi.org/10.1186/s12967-024-05068-z). Researchers compared the PDAC cell line PANC-1 with non-tumorigenic HPDE6c7 cells using an iodoTMT-based enrichment strategy followed by Orbitrap LC-MS/MS—an approach that mirrors our own Cysteine Profiling Service workflow.
| Project Snapshot | Outcome |
|---|---|
| Scope | Triplicate, label-based quantitation of reversible cysteine oxidation across two cell states |
| Depth | > 4,300 oxidized sites on > 2,100 proteins; 1,715 sites showed significant differential oxidation |
| Key Finding | Selective oxidation of HIF-1 pathway regulators—notably prolyl hydroxylase PHD2—suggested that redox control, rather than expression level, drives hypoxia signaling in PDAC |
| Drug-Discovery Relevance | Oxidation-sensitive cysteines clustered around HIF-1 regulatory nodes, flagging potential covalent-allosteric sites for small-molecule intervention and providing a mechanistic read-out for evaluating redox-modulating chemotypes |
Target Validation: Oxidation hotspots on PHD2 and VHL adaptor proteins pointed to redox gating of HIF-1α degradation—an actionable vulnerability for hypoxia-targeted therapies.
Biomarker Development: Cysteine-specific oxidation ratios served as pharmacodynamic markers to track the efficacy of emerging HIF-1 inhibitors in follow-up compound screens.
Combination Concepts: Overlaying cysteine oxidation changes with phosphoproteomic data suggested synergistic inhibition of both redox buffering and kinase-signaling arms, guiding rational drug-combination design.
This study illustrates how deep-scale cysteine PTM profiling can surface functional drug-response markers that would remain invisible to transcriptomics or total-proteome surveys alone.
Publication
Here are some publications in PTMs Proteomics research from our clients:

- Regulation of cardiac ferroptosis in diabetic human heart failure: uncovering molecular pathways and key targets. 2024. Cell Death Discovery. https://doi.org/10.1038/s41420-024-02044-w
- HDAC4 influences the DNA damage response and counteracts senescence by assembling with HDAC1/HDAC2 to control H2BK120 acetylation and homology-directed repair. Nucleic Acids Research. 2024. https://doi.org/10.1093/nar/gkae501
- Synergistic Combined-proteomics Guided Mapping strategy identifies mTOR mediated phosphorylation of LARP1 in nutrient responsiveness and dilated cardiomyopathy. BioRxiv. 2022. https://doi.org/10.1101/2022.10.13.512080
- In Vitro Identification of Phosphorylation Sites on TcPolβ by Protein Kinases TcCK1, TcCK2, TcAUK1, and TcPKC1 and Effect of Phorbol Ester on Activation by TcPKC of TcPolβ in Trypanosoma cruzi Epimastigotes. Microorganisms. 2024. https://doi.org/10.3390/microorganisms12050907
References
- Yanzheng Meng, Lin Zhang, Laizhi Zhang, Ziyu Wang, Xuanwen Wang, Chan Li, Yu Chen, Shipeng Shang, Lei Li, CysModDB: a comprehensive platform with the integration of manually curated resources and analysis tools for cysteine posttranslational modifications, Briefings in Bioinformatics, 2022;, bbac460, https://doi.org/10.1093/bib/bbac460
- White MEH, Gil J, Tate EW. Proteome-wide structural analysis identifies warhead- and coverage-specific biases in cysteine-focused chemoproteomics. Cell Chem Biol. 2023 Jul 20;30(7):828-838.e4. DOI: 10.1016/j.chembiol.2023.06.021
- Biggs, G.S., Cawood, E.E., Vuorinen, A. et al. Robust proteome profiling of cysteine-reactive fragments using label-free chemoproteomics. Nat Commun 16, 73 (2025). https://doi.org/10.1038/s41467-024-55057-5
- Boatner LM, Palafox MF, Schweppe DK, Backus KM. CysDB: a human cysteine database based on experimental quantitative chemoproteomics. Cell Chem Biol. 2023 Jun 15;30(6):683-698.e3. https://pubmed.ncbi.nlm.nih.gov/37119813/
- Yan T, Palmer AB, Geiszler DJ, Polasky DA, Boatner LM, Burton NR, Armenta E, Nesvizhskii AI, Backus KM. Enhancing Cysteine Chemoproteomic Coverage through Systematic Assessment of Click Chemistry Product Fragmentation. Anal Chem. 2022 Mar 8;94(9):3800-3810. DOI: 10.1021/acs.analchem.1c04402

















