Zeatin Riboside Analysis Service
Creative Proteomics provides ultra-sensitive LC-MS/MS analysis of Zeatin Riboside and related metabolites, enabling accurate quantification across various sample types. Our service supports RNA modification research, biomarker discovery, and pathway analysis, helping scientists decode metabolic regulation and disease mechanisms with precision and reproducibility.
Submit Your Request Now
×- What We Provide
- Advantage
- Workflow
- Technology Platform
- Sample Requirements
- FAQ
- Publications
What is Zeatin Riboside?
Zeatin Riboside (ZR) is a biologically active cytokinin, a class of adenine derivatives that regulate a myriad of plant physiological processes. As a riboside conjugate of zeatin, ZR is a key transport and storage form of cytokinin in plants. It influences processes including shoot and root development, apical dominance, senescence delay, and nutrient mobilization.
Molecular Formula: C15H23N5O5
Molecular Weight: 353.37 g/mol
ZR is particularly notable for its systemic mobility, allowing it to mediate hormone signaling between root and shoot tissues. Monitoring its concentration and metabolism provides critical insights into developmental regulation and stress responses in plants.
Why Analyze Zeatin Riboside?
Zeatin riboside analysis is critical for:
- Studying growth regulation: ZR influences apical dominance, morphogenesis, and metabolic activity.
- Optimizing agricultural practices: Quantifying ZR levels aids in improving crop yields and stress resilience.
- Biotechnological applications: Monitoring ZR in cell cultures helps optimize tissue differentiation (e.g., shoot vs. root formation). Accurate analysis enables researchers to correlate ZR fluctuations with phenotypic changes, advancing plant biology and agronomy.
Zeatin Riboside Analysis Service Offered by Creative Proteomics
- Quantitative Zeatin Riboside Measurement: We offer precise quantification of zeatin riboside using isotope-labeled internal standards and LC-MS/MS, enabling sensitive detection across various plant tissues.
- Full Cytokinin Profiling: This service includes simultaneous measurement of key cytokinins such as trans-zeatin, cis-zeatin, isopentenyladenine (iP), and their riboside and glucoside derivatives.
- Metabolic Pathway Analysis: We detect biosynthetic and degradation intermediates related to Zeatin Riboside, allowing in-depth understanding of cytokinin metabolism.
- Tissue-Specific Distribution Analysis: We analyze and compare zeatin riboside levels across different tissues, such as leaf, root, stem, seed, and flower.
- Time-Course Studies: This service tracks changes in zeatin riboside levels over time or under specific treatments, useful for developmental and stress-response research.
- Custom Method Development: We develop and optimize detection methods for unique species, sample types, or specific cytokinin analogs.
- Hormone Interaction Analysis: Zeatin riboside analysis can be combined with other hormone data (e.g., auxin, ABA, gibberellins) to study signaling interactions.
- Metabolic Flux Studies: Using stable isotope-labeled compounds, we measure biosynthesis and degradation rates of zeatin riboside in real time.
- Stress Condition Profiling: We quantify zeatin riboside levels under abiotic and biotic stress conditions to support research into plant adaptation and resilience.
List of Detected Zeatin Riboside and Related Metabolites
| Category | Analyte Name | Biological Role / Pathway |
|---|---|---|
| Primary Target | Zeatin Riboside (i⁶A) | tRNA modification; translation regulation |
| Upstream Precursors | Isopentenyl Pyrophosphate (IPP) | Mevalonate pathway; cytokinin biosynthesis |
| Dimethylallyl Pyrophosphate (DMAPP) | Precursor for prenylation reactions | |
| AMP (Adenosine Monophosphate) | Purine metabolism | |
| tRNA with unmodified adenosine (A37) | Precursor to i⁶A modification | |
| Related Modified Nucleosides | 2-Methylthio-Zeatin Riboside (ms²i⁶A) | Modified form in mitochondrial/archaeal tRNAs |
| N6-Zeatin Riboside Monophosphate | Intermediate in enzymatic transfer pathways | |
| Byproducts / Degradation | Adenosine | Nucleoside metabolism |
| Inosine | Result of A-to-I RNA editing | |
| Hypoxanthine | Purine degradation product | |
| Associated Pathways | Cytokinin Ribosides (e.g., Isopentenyladenine Riboside) | Plant hormone signaling |
| Methylthioadenosine (MTA) | Linked to polyamine and purine salvage pathways | |
| S-Adenosylmethionine (SAM) | Methyl donor for tRNA modification |
Advantages of Zeatin Riboside Assay
- Ultra-high Sensitivity: Detection limits as low as 0.1 ng/mL, enabling trace-level cytokinin profiling.
- Quantitative Accuracy: Standard curves with R² > 0.995 ensure highly reproducible results.
- Multi-matrix Compatibility: Validated for over 30 plant tissues and fluids, including leaf, root, seed, sap.
- High Throughput: Capable of processing 200+ samples per batch, ideal for large-scale screening.
- Comprehensive Coverage: Detects over 40 cytokinin-related metabolites, providing a complete metabolic view.
- Turnaround Time: Industry-leading speed with standard 14–28 working days delivery.
Workflow for Zeatin Riboside Analysis Service
Technology Platform for Zeatin Riboside Analysis Service
Primary Platform: LC-MS/MS using Sciex QTRAP 6500+ offers femtomole-level sensitivity and excellent reproducibility.
Chromatographic System: UHPLC system (Agilent 1290 Infinity II) equipped with C18 reversed-phase columns for sharp peak resolution.

SCIEX Triple Quad™ 6500+ (Figure from Sciex)

Agilent 1260 Infinity II HPLC (Fig from Agilent)
Sample Requirements for Zeatin Riboside Analysis Service
| Sample Type | Minimum Quantity | Storage Condition |
|---|---|---|
| Fresh Plant Tissue (leaf, root, shoot) | ≥ 0.5 g | Liquid nitrogen or −80°C |
| Freeze-dried Tissue | ≥ 50 mg | Room temp or −20°C |
| Plant Sap or Xylem Exudate | ≥ 100 μL | −80°C |
| Culture Media or Hydroponic Solution | ≥ 1 mL | −20°C |
| Seed Samples | ≥ 0.3 g | −20°C |
Applications of Zeatin Riboside Assay Service
Epitranscriptomics Research
Investigating RNA modifications and their roles in gene regulation and translational control.
Plant Physiology
Studying cytokinin biosynthesis pathways and hormone-regulated growth responses.
Functional Genomics
Exploring the impact of tRNA modifications on protein expression and codon usage bias.
Metabolic Flux Analysis
Tracing purine metabolism and nucleoside turnover under varying nutrient or stress conditions.
Synthetic Biology
Engineering pathways involving prenylated nucleosides for expanded genetic coding systems.
Microbial Adaptation Studies
Analyzing RNA modification patterns in prokaryotes under environmental or antibiotic stress.
FAQ of Zeatin Riboside Analysis Service
What is the recommended storage condition for samples before sending them for Zeatin Riboside analysis?
Samples should be flash-frozen immediately after collection and stored at -80°C. Avoid repeated freeze-thaw cycles to preserve analyte stability.
Can I submit freeze-dried plant tissue for analysis?
Yes, freeze-dried (lyophilized) samples are acceptable. Please ensure samples are well-labeled and stored in airtight containers to prevent moisture absorption.
What is the typical turnaround time for Zeatin Riboside analysis results?
Standard turnaround is 14–28 business days from sample receipt, depending on sample volume and project complexity. Expedited services may be available on request.
Is it possible to analyze Zeatin Riboside in both free and conjugated forms?
Yes, we can customize the method to differentiate between free and conjugated (glucosylated or riboside-bound) forms of Zeatin upon request.
How should I ship my samples for Zeatin Riboside analysis?
Ship samples on dry ice in a well-insulated container. Include a detailed sample list and contact us in advance to schedule delivery and ensure proper handling.
Can your analysis detect other cytokinin types along with Zeatin Riboside?
Yes, we offer multiplex cytokinin profiling that includes zeatin, zeatin riboside, kinetin, isopentenyladenine, and related metabolites.
Do I need to perform any extraction or preparation before sending plant samples?
No pre-treatment is required. We recommend sending raw, frozen material. Our team will perform optimized extraction and purification protocols in-house.
Learn about other Q&A.
Zeatin Riboside Analysis Service Case Study

Title: A high trans-zeatin nucleoside concentration in corms may promote the multileaf growth of Amorphophallus muelleri
Journal: Frontiers in Plant Science
Published: 2024
- Background
- Materials & Methods
- Results
Amorphophallus muelleri, a pearl-bud-type konjac, exhibits a unique multileaf growth pattern, unlike the single-leaf-per-season growth seen in most konjac species like A. konjac. This sequential leaf emergence enhances corm development and glucomannan yield, offering significant agronomic and economic benefits. However, the hormonal mechanisms governing this trait remain unclear. The study focuses on the dynamics of plant hormones—particularly cytokinins (notably trans-zeatin riboside, tZR), indole-3-acetic acid (IAA), and abscisic acid (ABA)—during different stages of leaf sprouting. It investigates how shifts in hormone concentrations and related gene expressions in corms correlate with axillary bud activation and multileaf development in A. muelleri.
Plant Growth and Sampling:
Two-year-old A. muelleri corms (~100 g) were cultivated in 10 L pots in a greenhouse (Kunming University, China) from May to October 2021 under 25–35°C, 50% shade, and 60–80% humidity. Developmental stages sampled included dormancy, sprouting, first and second leaf maturation, and lodging. Leaves and corm tissues were collected accordingly, with three biological replicates per stage.
Endogenous Phytohormone Analysis:
Phytohormones were extracted from frozen samples using acetonitrile-based solution with isotopically labeled standards. After homogenization and centrifugation, samples were filtered and analyzed using UHPLC-MS/MS (Waters ACQUITY UPLC with SCIEX systems). Calibration curves ensured quantification accuracy; precision was confirmed via RSD and recovery rates.
RNA-Seq Analysis:
RNA was extracted from corm tissues at four growth stages and sequenced using Illumina NovaSeq6000. Clean reads were aligned to the konjac reference genome (PRJNA608095) using HISAT2, and transcript abundance was quantified by FPKM via StringTie and RSEM. Differential expression was defined as adjusted P < 0.05 and fold change >2.
Exogenous iP Application:
Dormant corms (~5 g) were treated with 50 or 100 mg L⁻¹ iP for 3 h, then grown under controlled conditions (28°C, 16/8 h photoperiod). After 30 days, growth parameters (bud/root number and length) were measured. Hormone levels were analyzed in treated tissues.
Statistical Analysis:
One-way ANOVA and Tukey's test (P ≤ 0.05) were applied using SPSS 16.0. Heatmaps and plots were created with TBtools and SigmaPlot 10.0.
Cytokinin Dynamics:
Five cytokinins (KT, tZR, tZ, iP, iPA) showed stage-specific changes. In corms, KT, tZR, and tZ increased during early growth, with tZR peaking sharply (>5000× dormant level) at the second leaf sprouting stage. iP and iPA peaked in dormancy and decreased afterward. Leaf cytokinin levels were lower overall, with KT being the most abundant.
Effects of Exogenous iP:
Application of 100 mg L⁻¹ iP promoted bud/root growth and increased multibud formation. iP levels in buds were 30× higher than in corms under treatment, making up ~93% of total plant content. tZR also increased in buds. iP elevated IAA in corms and roots but reduced it in buds at low concentration. ABA decreased in buds and roots but rose in corms.
Gene Expression in Cytokinin Pathways:
IPT3/5 were upregulated in the second leaf mature stage. CYP735A1/2 showed strong induction during the second leaf sprouting stage. LOG expression rose moderately. CKX3/5/6 and UGT75L6, involved in cytokinin degradation, declined as the second leaf sprouted.
IAA and ABA Patterns:
IAA peaked in corms at the second leaf sprouting stage, correlating with high AO1 and PIN1A expression. ABA was highest in dormant corms and dropped sharply with bud emergence. NCED1 expression decreased, while ABA-degrading genes (CCD, PP2C51) increased during sprouting.
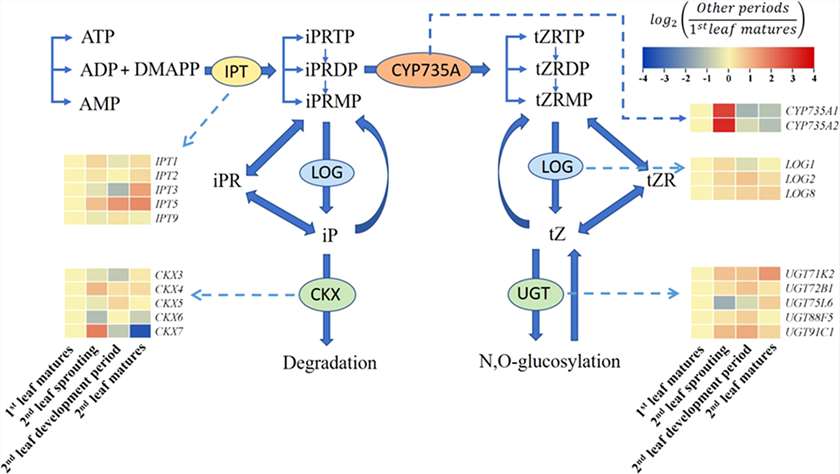 Summary of the cytokinin biosynthesis and catabolism pathways.
Summary of the cytokinin biosynthesis and catabolism pathways.
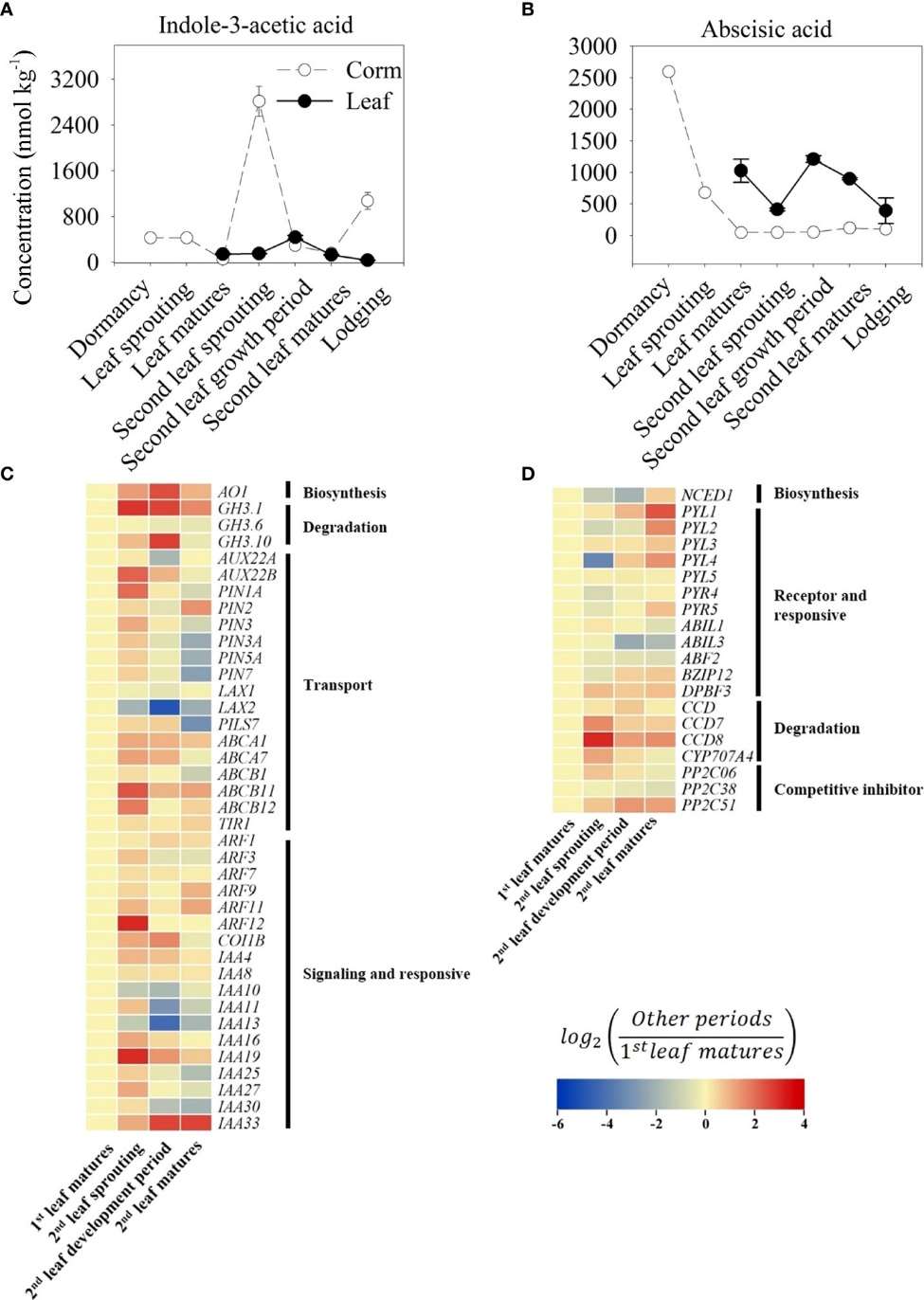 Trends of indole-3-acetic acid (IAA, A) and abscisic acid (ABA, B) content in growth cycle of A. muelleri.
Trends of indole-3-acetic acid (IAA, A) and abscisic acid (ABA, B) content in growth cycle of A. muelleri.
Reference
- Xue, Zhiqin, et al. "A high trans-zeatin nucleoside concentration in corms may promote the multileaf growth of Amorphophallus muelleri." Frontiers in Plant Science 13 (2022): 964003. https://doi.org/10.3389/fpls.2022.964003
Publications
Here are some publications in Metabolomics research from our clients:

- The Suppression of the KRAS G12D-Nrf2 Axis Shifts Arginine into the Phosphocreatine Energy System in Pancreatic Cancer Cells. 2023. https://doi.org/10.1016/j.isci.2023.108566
- The Brain Metabolome Is Modified by Obesity in a Sex-Dependent Manner. 2024. https://doi.org/10.3390/ijms25063475
- Lipin-1 regulates lipid catabolism in pro-resolving macrophages. 2020. https://doi.org/10.1101/2020.06.03.121293
- Pan-lysyl oxidase inhibition disrupts fibroinflammatory tumor stroma, rendering cholangiocarcinoma susceptible to chemotherapy. 2024. https://doi.org/10.1097/HC9.0000000000000502
- Neddylation is required for perinatal cardiac development through stimulation of metabolic maturation. 2023. https://doi.org/10.1016/j.celrep.2023.112018







