Shikimic Acid Analysis Service
Creative Proteomics provides high-sensitivity shikimic acid analysis through targeted quantification and broad pathway metabolite profiling using LC-MS/MS, HPLC, and GC-MS.
We help you:
- Achieve precise quantification with LOD as low as 1 ng/mL
- Monitor over 10 key metabolites in the shikimate pathway
- Visualize metabolic flux with integrated KEGG-based pathway mapping
Optimized for plant tissues, microbial cultures, and fermentation samples—accelerate your metabolic research and biotechnological applications with reliable, high-throughput data.
Submit Your Request Now
×- What We Provide
- Advantage
- Workflow
- Technology Platform
- Sample Requirements
- FAQ
- Case Study
Overview of Shikimic Acid
Shikimic acid is a pivotal biochemical intermediate in the shikimate pathway, responsible for the biosynthesis of aromatic amino acids and various secondary metabolites in microorganisms and plants. This pathway does not occur in animals, making its metabolites highly relevant in agriculture, biotechnology, and synthetic biology. Quantitative and qualitative analysis of shikimic acid provides critical insights into metabolic flux, strain engineering, herbicide efficacy, and fermentation optimization.
Why Analyze Shikimic Acid?
Shikimic acid is a critical intermediate in the shikimate pathway, a metabolic route present in bacteria, fungi, and plants but absent in mammals. This unique characteristic makes it highly relevant in both basic and applied life sciences. Accurate quantification and profiling of shikimic acid supports a range of scientific investigations and biotechnological applications:
- Metabolic Engineering: Evaluate the effects of genetic modifications on flux through the shikimate pathway in microbial or plant systems.
- Herbicide Mechanism Studies: Monitor metabolic disturbances caused by inhibitors targeting enzymes such as EPSP synthase.
- Synthetic Biology Optimization: Measure pathway intermediates in engineered strains to improve biosynthesis of aromatic compounds.
- Plant Stress Response Research: Investigate changes in shikimate metabolism under biotic or abiotic stress conditions.
- Nutritional and Agricultural Studies: Characterize plant metabolic profiles for crop quality, resistance, and yield research.
Shikimic Acid Analysis Service Offered by Creative Proteomics
- Shikimic Acid Quantification: Precise measurement of shikimic acid using isotope-dilution LC-MS/MS with high sensitivity and specificity.
- Pathway Intermediate Profiling: Detection of upstream and downstream metabolites, including 3-dehydroshikimate, chorismate, and prephenate.
- Targeted Metabolite Panels: Custom-designed panels tailored to plant, microbial, or engineered systems for shikimate pathway analysis.
- Kinetic/Time-Course Analysis: Monitoring of metabolite dynamics across multiple time points to support flux and fermentation studies.
- Comparative Condition Analysis: Quantitative comparisons between experimental conditions, treatments, or engineered strains.
- Bioinformatics & Pathway Mapping: Visualization of metabolite changes in KEGG/MetaCyc pathways, with integrated statistical analysis.
- Custom Analytical Support: Flexible, project-specific method development with expert scientific consultation.
Detected Shikimic Acid and Related Analytes
| Compound Name | Chemical Class | Associated Pathway | Biological Role |
|---|---|---|---|
| Shikimic Acid | Cyclohexene Tricarboxylic Acid | Shikimate Pathway | Central intermediate in the biosynthesis of aromatic compounds |
| 3-Dehydroshikimic Acid | Keto Acid | Shikimate Pathway | Precursor to shikimic acid and quinic acid |
| Quinic Acid | Cyclitol | Secondary Metabolism | Related to chlorogenic acid synthesis in plants |
| 3-Phosphoshikimic Acid | Phosphorylated Sugar Acid | Shikimate Pathway | Key intermediate between shikimate and chorismate |
| Chorismic Acid (Chorismate) | Aromatic Acid Precursor | Aromatic Amino Acid Biosynthesis | Branch point for synthesis of tryptophan, phenylalanine, tyrosine |
| Prephenic Acid (Prephenate) | Aromatic Acid Derivative | Phenylalanine/Tyrosine Biosynthesis | Intermediate in synthesis of phenylalanine and tyrosine |
| Anthranilic Acid | Aromatic Amine | Tryptophan Biosynthesis | Early intermediate toward tryptophan |
| Phenylpyruvic Acid | Keto Acid | Phenylpropanoid Pathway | Derived from phenylalanine; precursor to flavonoids |
| Tyrosine | Aromatic Amino Acid | Protein and Secondary Metabolism | End product of the shikimate-derived branch |
| Phenylalanine | Aromatic Amino Acid | Protein and Phenylpropanoid Pathway | Substrate for flavonoid and lignin biosynthesis |
| Tryptophan | Aromatic Amino Acid | Indole Alkaloid and Protein Synthesis | Final product of the chorismate-derived branch |
Advantages of Shikimic Acid Assay
- High Sensitivity: Limit of detection (LOD) as low as 1 ng/mL for shikimic acid
- Excellent Recovery Rate: Average recovery of 92.5% ± 3.1% across plant and microbial matrices
- High Precision: Intra-assay coefficient of variation (CV) < 5% in technical replicates
- Wide Dynamic Range: Linear range covering 4–5 orders of magnitude (R² > 0.995)
- Robust Throughput: Capable of processing 96 samples per run using automated LC-MS/MS
- Comprehensive Metabolite Coverage: Detection of over 10 key metabolites in the shikimate pathway
- Integrated Statistical: Analysis PCA, hierarchical clustering, and pathway enrichment included
- Custom Method Flexibility: Parameterizable sample prep and detection modes tailored to study design
- Data Deliverables: Raw data + fully quantified reports + annotated metabolic pathway diagrams
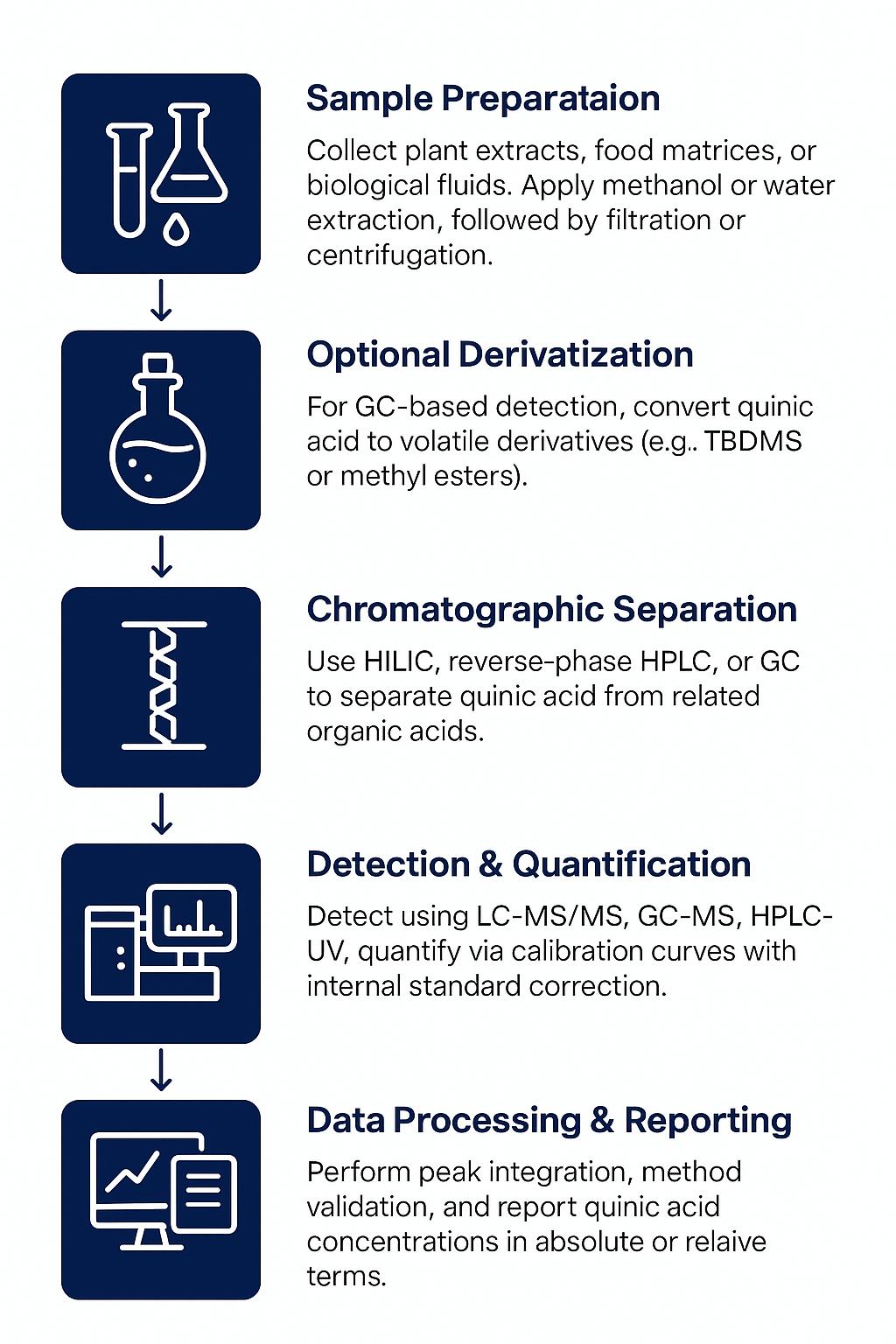
Workflow for Shikimic Acid Analysis Service
Technology Platform for Shikimic Acid Analysis Service
Agilent 6495C Triple Quadrupole LC-MS/MS: Used for high-sensitivity targeted quantification of shikimic acid and pathway intermediates. Achieves detection limits as low as 1 ng/mL with excellent precision and selectivity.
Agilent 1260 Infinity II HPLC: Provides robust chromatographic separation with high resolution and reproducibility. Compatible with both polar and non-polar metabolites in various biological matrices.
Agilent 7890B GC + 5977A: Optional GC-MS analysis for volatile or derivatized compounds. Applied to specialized matrices such as processed plant or microbial fermentation samples.

Agilent 6495C Triple Quadrupole (Figure from Agilent)

Agilent 7890B-5977A (Figure from Agilent)

Agilent 1260 Infinity II HPLC (Fig from Agilent)
Sample Requirements for Shikimic Acid Analysis Service
| Sample Type | Required Volume / Weight | Notes |
|---|---|---|
| Plant Tissue (leaf, root, stem) | ≥ 200 mg (fresh weight) | Preferably snap-frozen in liquid nitrogen and stored at –80°C |
| Microbial Pellets | ≥ 100 mg (wet weight) | Harvest during logarithmic phase; freeze-dried or frozen |
| Cell Cultures / Suspension Cells | ≥ 1 × 10⁷ cells or ≥ 500 µL | Centrifuge to pellet; wash with PBS before freezing |
| Fermentation Broth / Supernatant | ≥ 1 mL | Filtered or centrifuged to remove solids; store at –20°C or below |
| Serum / Plasma | ≥ 200 µL | Collected with anticoagulant; avoid repeated freeze-thaw cycles |
| Urine / Plant Exudates | ≥ 500 µL | Collected fresh; filter to remove particulates |
FAQ of Shikimic Acid Analysis Service
Can you analyze shikimic acid in beverages like wine?
Yes, we specialize in detecting shikimic acid in complex matrices such as wine, where it serves as a marker for grape variety differentiation. Our protocols include matrix-specific cleanup steps to eliminate interferents like tannins and sugars.
Is there a recommended method for stabilizing shikimic acid in plant tissues before extraction?
Samples should be snap-frozen in liquid nitrogen immediately after collection to prevent enzymatic degradation. For long-term storage, maintain tissues at –80°C in airtight containers to avoid moisture absorption .
Do you provide isotopic labeling studies for metabolic flux analysis?
Yes, we offer custom stable isotope (e.g., ¹³C-labeled) tracer experiments to quantify shikimate pathway dynamics in real time, enabling precise tracking of metabolic intermediates.
How do you handle samples with low shikimic acid concentrations (e.g.,<1 ng/mL)?
For trace-level detection, we utilize LC-MS/MS with pre-concentration techniques, achieving sensitivity down to 0.5 ng/mL by optimizing solid-phase extraction (SPE) protocols.
Can your service distinguish shikimic acid isomers like quinic acid?
Absolutely. Our HPLC and LC-MS methods resolve structurally similar compounds (e.g., quinic acid) using gradient elution and selective ion monitoring, ensuring >99% specificity.
What bioinformatics tools do you use for pathway mapping?
We integrate KEGG and MetaCyc databases with in-house algorithms to visualize metabolite changes, identify rate-limiting steps, and correlate shikimate flux with downstream aromatic compound synthesis.
Do you support analysis of transgenic microbial strains overproducing shikimic acid?
Yes. We validate engineered strains (e.g., E. coli hyperproducers) using comparative profiling and kinetic studies, with protocols optimized for high-yield fermentation broths.
Learn about other Q&A.
Shikimic Acid Analysis Service Case Study

Title: The Analysis of Leontopodium leontopodioides (Willd.) Beauv. Chemical Composition by GC/MS and UPLC-Q-Orbitrap MS
Journal: International Journal of Analytical Chemistry
Published: 2024
- Background
- Methods
- Results
- Reference
Leontopodium leontopodioides (Willd.) Beauv., a traditional Chinese medicinal herb used primarily for treating lung-related conditions, had not undergone a comprehensive chemical composition study prior to this research. While previous studies identified select compounds—mainly flavonoids and phenolic acids—using conventional chromatographic techniques, a full-spectrum chemical profile of both water extracts and essential oils remained unexplored. This study systematically analyzes the phytochemical constituents using advanced UPLC-Q-Orbitrap MS and GC-MS techniques, identifying 39 compounds in water extracts and 17–18 volatile components in essential oils, with 18 of these compounds newly reported in this species. The findings offer a foundational chemical characterization for future pharmacodynamic research and quality evaluation.
The study employed UPLC-Q-Orbitrap MS and GC-MS to comprehensively characterize the chemical composition of Leontopodium leontopodioides.
Sample Preparation:
Water extract was prepared via triple water decoction, yielding 27.63%. Essential oil was obtained by supercritical CO₂ extraction at 45°C and 18 MPa for 5 hours, yielding 1.20 mL of oil.
UPLC-Q-Orbitrap MS Analysis:
Samples were dissolved in 80% methanol, ultrasonicated, centrifuged, and injected for analysis. Chromatographic separation was performed on a Waters HSS T3 C18 column using a gradient of 0.1% formic acid in water/acetonitrile. Detection employed HESI in both positive and negative ion modes with full MS/dd-MS² scanning. Compound Discoverer 3.2, mzCloud, and mzVault were used for identification.
GC-MS Analysis:
Essential oil samples dissolved in n-hexane or diethyl ether were analyzed using an HP-5 ms column. The oven was programmed across multiple temperature ramps with helium as the carrier gas. EI mode at 70 eV was used for ionization, scanning m/z 35–450. Compound identification was based on NIST library matching and peak area normalization.
Creative Proteomics Can Support This Research by Providing the Following Shikimic Acid-Focused Services:
Based on this workflow, Creative Proteomics can provide the following shikimic acid-related analytical services:
- Targeted LC-MS/MS quantification of shikimic acid and downstream phenolic acid derivatives (e.g., caffeic, ferulic, chlorogenic acids)
- Untargeted metabolomics for pathway mapping of the shikimate and phenylpropanoid biosynthesis
- GC-MS analysis of volatile and semi-volatile compounds derived from aromatic precursors
- Custom extraction optimization for plant matrices (e.g., decoction, supercritical CO₂, methanol-based)
- Data interpretation support with KEGG-based pathway integration and compound identification via in-house and public spectral libraries
Chemical Composition of L. leontopodioides Water Extract (UPLC-Q-Orbitrap MS)
A total of 39 compounds were identified, including:
- Flavonoids and Glycosides (13 compounds): Luteolin, cynaroside, and astragalin were identified based on characteristic fragment ions and database matching.
- Phenolic Acids (15 compounds): Notable examples include protocatechuic acid and shikimic acid, identified via typical CO₂ and H₂O losses.
- Pentacyclic Triterpenoids (4 compounds): Ursolic acid was confirmed by stepwise fragmentation involving H₂O and COOH loss.
- Oligosaccharides and Glycosides (4 compounds): Identified by glycosidic bond cleavage; sucrose was a representative compound.
- Other Compounds: Two alkaloids (e.g., piperine) and one coumarin were identified.
Volatile Components of L. leontopodioides Essential Oil (GC-MS)
33 volatile compounds were identified from n-hexane and diethyl ether extracts (7 shared):
- n-Hexane Extract: Dominant: phenylpropenes (64.52%), monoterpenes (10.96%), fatty acids (10.03%).
Main compounds: methylconiferylaldehyde (14.77%), (E)-2,6-dimethoxy-4-(prop-1-en-1-yl) phenol (12.04%), eugenol (11.51%). - Diethyl Ether Extract: Dominant: fatty acids (28.99%), phenylpropenes (28.37%).
Main compounds: methylconiferylaldehyde (9.61%), pentadecanoic acid (9.25%), 8-methylnonanoic acid (8.63%). - Common Components: Terpinolene, terpinen-4-ol, γ-terpinene, methyleugenol, methylconiferylaldehyde, tetradecanoic acid, n-hexadecanoic acid.
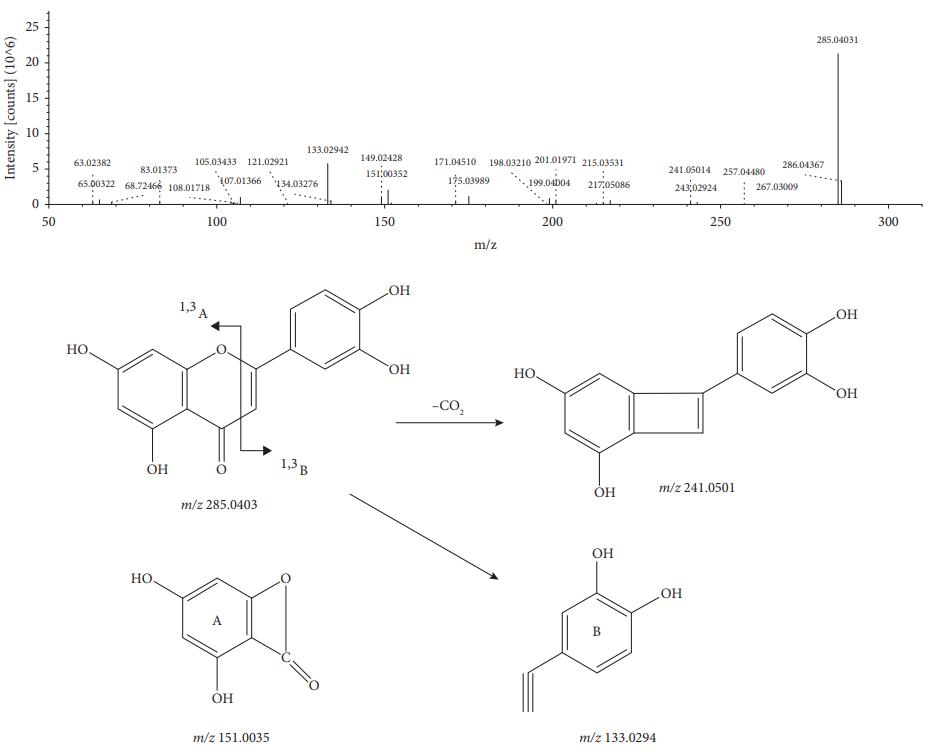 MS/MS spectrum and the fragmentation pathway of luteolin.
MS/MS spectrum and the fragmentation pathway of luteolin.
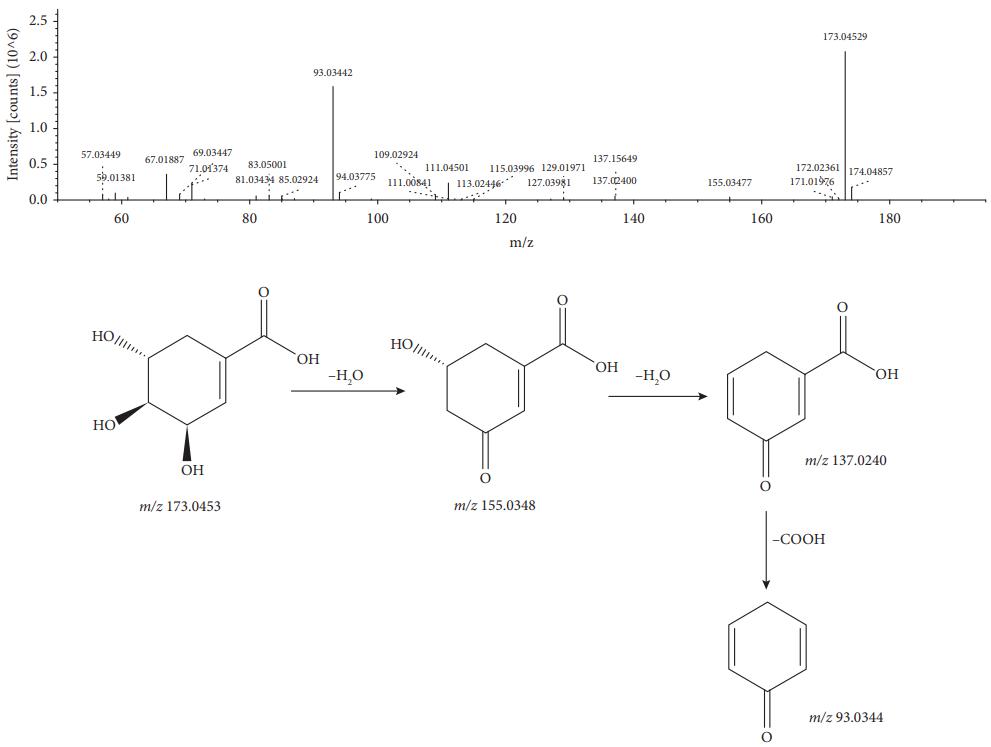 MS/MS spectrum and the fragmentation pathway of shikimic acid.
MS/MS spectrum and the fragmentation pathway of shikimic acid.
Reference
- Chen, Yuanyuan, et al. "The Analysis of Leontopodium leontopodioides (Willd.) Beauv. Chemical Composition by GC/MS and UPLC‐Q‐Orbitrap MS." International Journal of Analytical Chemistry 2024.1 (2024): 3525212. https://doi.org/10.1155/2024/3525212
Publications
Here are some publications in Metabolomics research from our clients:

- Disruption of CYCLOPHILIN 38 function reveals a photosynthesis-dependent systemic signal controlling lateral root emergence. 2020. https://doi.org/10.1101/2020.03.11.985820
- Comparative metabolite profiling of salt sensitive Oryza sativa and the halophytic wild rice Oryza coarctata under salt stress. 2024. https://doi.org/10.1002/pei3.10155
- Macrophage-associated lipin-1 promotes β-oxidation in response to proresolving stimuli. 2020. https://doi.org/10.4049/immunohorizons.2000047
- Overexpression of maize ZmLOX6 in Arabidopsis thaliana enhances damage-induced pentyl leaf volatile emissions that affect plant growth and interaction with aphids. 2023. https://doi.org/10.1093/jxb/erac522
- Untargeted metabolomics reveal sex-specific and non-specific redox-modulating metabolites in kidneys following binge drinking. 2023. https://doi.org/10.1530/REM-23-0005






