Malonic Acid Analysis Service
Creative Proteomics offers high-sensitivity malonic acid analysis through targeted quantification and pathway-level profiling using LC-MS/MS, HPLC, and GC-MS.
We help you:
- Achieve precise quantification with detection limits as low as 1 ng/mL
- Monitor 15+ related metabolites across the TCA cycle and dicarboxylic acid metabolism
- Map carbon flux and mitochondrial activity with integrated metabolic pathway analysis
Optimized for plasma, microbial cultures, plant tissues, and environmental samples—accelerate your metabolic, bioengineering, or ecological research with reliable, high-throughput data.
Submit Your Request Now
×- What We Provide
- Advantage
- Workflow
- Technology Platform
- Sample Requirements
- FAQs
- Case Study
Overview of Malonic Acid
Malonic acid (propanedioic acid) is a dicarboxylic acid that plays a crucial intermediary role in various metabolic pathways, including the tricarboxylic acid (TCA) cycle and fatty acid biosynthesis. It is a known inhibitor of succinate dehydrogenase (SDH), thereby impacting mitochondrial respiration and energy metabolism. Elevated or dysregulated malonic acid levels are associated with metabolic disorders, mitochondrial diseases, and oxidative stress.
Why Analyze Malonic Acid?
Malonic acid is a key intermediate in several central metabolic pathways, including fatty acid biosynthesis and the TCA cycle. Its analysis provides valuable insights into cellular energy metabolism, carbon flux, and enzymatic regulation.
Key reasons to analyze malonic acid:
- Monitor Metabolic Flux: Track carbon flow in biosynthetic pathways such as fatty acid elongation.
- Assess Mitochondrial Function: As an SDH inhibitor, malonic acid reflects changes in mitochondrial activity and oxidative metabolism.
- Study Lipogenesis: Measure precursor availability for fatty acid synthesis in microbial, plant, or animal systems.
- Support Environmental Research: Evaluate microbial activity and nutrient cycling in ecological samples.
- Optimize Bioprocesses: Guide metabolic engineering in industrial fermentation and bioproduction systems.
Accurate quantification of malonic acid supports biochemical research, environmental monitoring, and industrial biotechnology.
Malonic Acid Analysis Service Offered by Creative Proteomics
- Quantitative Analysis of Malonic Acid: Absolute quantification in a variety of biological and environmental matrices using isotope-labeled internal standards.
- Targeted Metabolomics Profiling: Simultaneous detection of malonic acid and a panel of pathway-related metabolites involved in the TCA cycle, fatty acid metabolism, and dicarboxylic acid pathways.
- Pathway-Focused Data Interpretation: Mapping of malonic acid levels within metabolic networks to assess pathway activity and carbon flow distribution.
- Stable Isotope Tracing Analysis: Monitoring dynamic changes in labeled carbon incorporation into malonic acid for flux analysis studies.
- Comparative Metabolic Analysis: Multi-group comparison to identify malonic acid alterations under different experimental conditions or treatments.
- Custom Method Development: Development of tailored protocols for unique sample types or analytical needs, including unusual matrices or low-concentration targets.
- Batch Processing for High-Throughput Projects: Scalable analysis services for large sample cohorts in systems biology, microbial screening, or environmental monitoring.
- Raw Data and Statistical Reporting: Delivery of raw chromatograms, processed quantification data, and optional multivariate analysis or statistical summaries.
Detected Malonic Acid and Related Analytes
| Category | Detected Metabolites | Associated Metabolic Pathways |
|---|---|---|
| Core Analyte | Malonic Acid | Dicarboxylic Acid Metabolism, Fatty Acid Biosynthesis |
| TCA Cycle Intermediates | Succinic Acid, Fumaric Acid, Malic Acid, Citric Acid, α-Ketoglutarate | Tricarboxylic Acid (TCA) Cycle, Energy Metabolism |
| Fatty Acid Biosynthesis | Acetyl-CoA, Malonyl-CoA | Lipogenesis, Polyketide Synthesis |
| Amino Acid Metabolism | Glycine, Alanine, Glutamate, Serine | Glyoxylate and Dicarboxylate Metabolism, Nitrogen Cycle |
| Redox/Mitochondrial Markers | NAD⁺, NADH, Glutathione (GSH/GSSG), Succinate | Oxidative Phosphorylation, Mitochondrial Stress Response |
| Dicarboxylic Acids Panel | Oxalic Acid, Glutaric Acid, Methylmalonic Acid | Organic Acid Metabolism, Peroxisomal β-Oxidation |
Advantages of Malonic Acid Assay
- Ultra-High Sensitivity: Detection limits as low as 1–5 ng/mL using Agilent 6495C LC-MS/MS.
- Broad Quantification Range: Linear range from 5 ng/mL to 5000 ng/mL (R² > 0.995), covering diverse concentration levels.
- Excellent Precision: Intra-assay RSD<6%, inter-assay RSD <10%, ensuring stable, repeatable results.
- Versatile Sample Coverage: Validated for 10+ sample types including plasma, urine, tissues, and microbial cultures.
- High Throughput: Supports up to 96 samples/day, ideal for large-scale or time-critical projects.
- High Quantitative Accuracy: Internal standards and calibration strategies ensure<5% quantification error.
- Dual Platform Flexibility: LC-MS/MS and GC-MS platforms tailored to sample matrix and analyte properties.
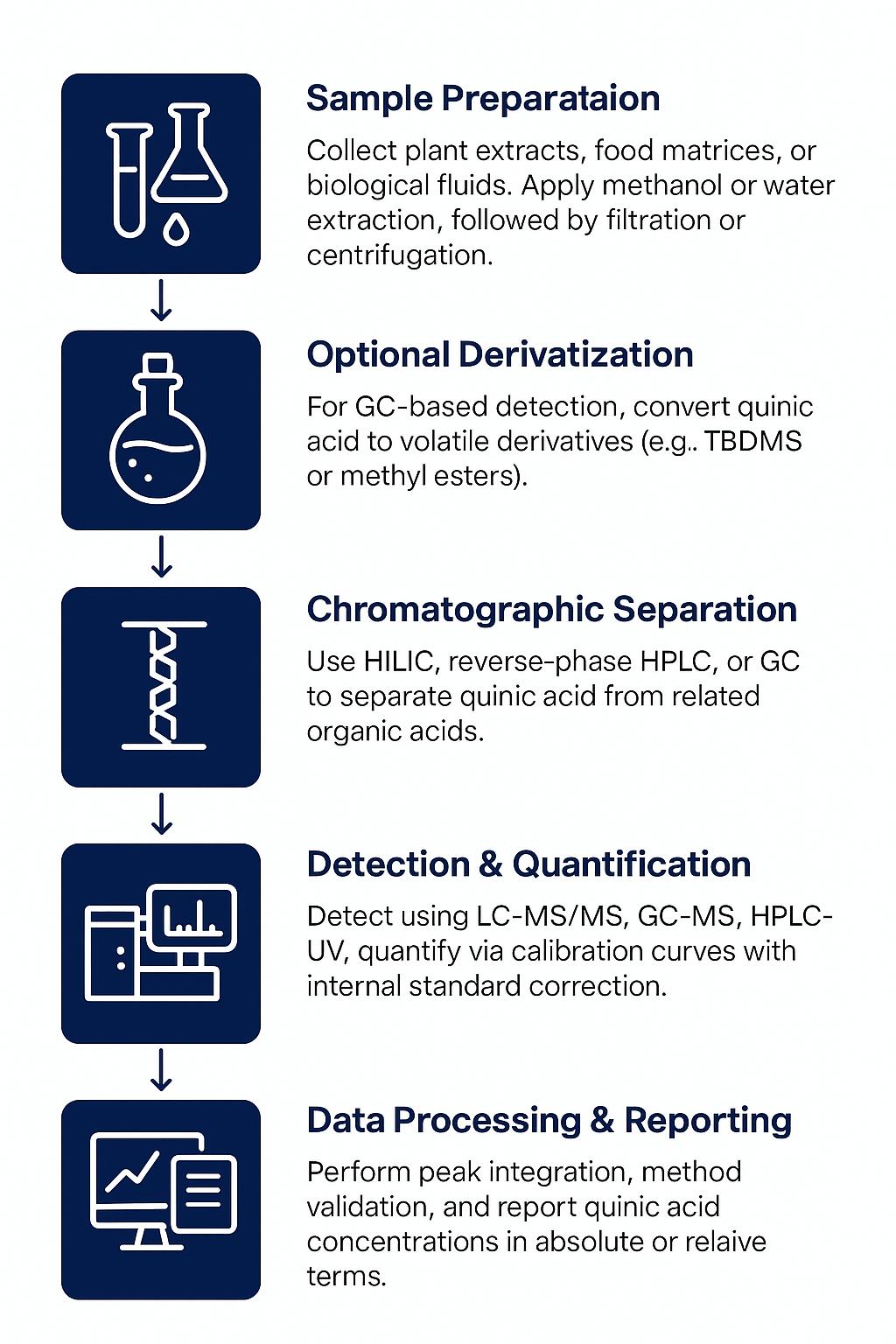
Workflow for Malonic Acid Analysis Service
Technology Platform for Malonic Acid Analysis Service
Agilent 6495C Triple Quadrupole LC-MS/MS: Utilized for high-sensitivity quantification in MRM mode, with femtogram-level detection and excellent selectivity in complex biological matrices.
Agilent 7890B GC with 5977A MSD: Suitable for derivatized malonic acid analysis in environmental and microbial samples. Offers high resolution and low background interference.
Agilent 1260 Infinity II HPLC: Provides robust chromatographic separation with high reproducibility, used in both LC-MS and UV workflows to enhance peak clarity and retention stability.

Agilent 6495C Triple Quadrupole (Figure from Agilent)

Agilent 7890B-5977A (Figure from Agilent)

Agilent 1260 Infinity II HPLC (Fig from Agilent)
Sample Requirements for Malonic Acid Analysis Service
| Sample Type | Required Volume / Weight | Notes |
|---|---|---|
| Plasma / Serum | ≥ 100 μL | Collected in EDTA or heparin tubes; avoid hemolysis |
| Urine | ≥ 200 μL | First morning urine preferred; no preservatives |
| Cell Pellet | ≥ 1 × 10⁶ cells | Washed with cold PBS; snap-frozen in liquid nitrogen |
| Tissue (animal/plant) | ≥ 20 mg (wet weight) | Flash-frozen immediately after collection; store at -80°C |
| Bacterial / Fungal Pellet | ≥ 50 mg (wet weight) | Harvest in log phase; wash to remove media |
| Culture Supernatant / Media | ≥ 500 μL | Filtered or centrifuged to remove particulates |
| Environmental Sample (Soil/Water) | ≥ 1 g (soil) / ≥ 500 μL (water) | Store at 4°C short-term or -20°C for long-term |
| CSF (Cerebrospinal Fluid) | ≥ 50 μL | Specialized request; please consult before submission |
FAQ of Malonic Acid Analysis Service
How should samples be stored and transported to ensure malonic acid stability?
For biological samples (e.g., plasma, tissues), immediate freezing at -80°C is recommended to prevent degradation. Ship samples on dry ice with temperature tracking. Microbial or cell culture samples should be quenched rapidly (e.g., liquid nitrogen) before extraction. Avoid repeated freeze-thaw cycles.
What is the turnaround time for malonic acid analysis?
Standard analysis typically takes 5-7 business days after sample receipt. Expedited services (3-4 days) are available for urgent projects, subject to instrument availability.
Can I integrate malonic acid data with other omics datasets (e.g., transcriptomics)?
Yes, we provide optional cross-omics integration support. Raw data files (e.g., .csv/.xlsx) are compatible with major bioinformatics tools, and we offer consultation for correlation analysis with gene expression or proteomics data.
How do you handle samples with very low malonic acid concentrations?
For trace-level detection, we employ pre-concentration techniques (e.g., lyophilization, solid-phase extraction) and ultra-sensitive LC-MS/MS protocols. Method validation for low-abundance samples is performed upon request.
Are there specific requirements for sample preparation in microbial cultures?
Yes, microbial cells should be harvested during log phase for accurate flux analysis. Include details on culture medium composition, as some components (e.g., carbon sources) may interfere with detection. We provide optimized extraction protocols for bacterial/fungal samples.
Can malonic acid levels be analyzed alongside other short-chain fatty acids?
Absolutely. Our dicarboxylic acids panel includes simultaneous quantification of malonic acid, succinic acid, and oxaloacetic acid. Custom panels can be designed to include specific short-chain fatty acids (e.g., acetic, propionic acid).
How are data normalized for variations in sample input (e.g., tissue weight differences)?
We use dual normalization strategies: (1) internal standard correction for technical variability and (2) biomolecule-based normalization (e.g., protein concentration, cell count) for biological variability. Clients can specify preferred normalization methods.
What quality control measures are included in the analysis?
Every batch includes blank controls, pooled QC samples, and reference standards. System suitability tests (retention time stability, peak shape) are performed daily. Full QC reports are provided with deliverables.
Can I request re-analysis if results appear inconsistent?
Yes, we offer free re-analysis for technical replicates if outliers are detected in QC metrics. Additional charges apply for new sample preparations or extended projects.
How are results presented for multi-group comparative studies?
Data are delivered with fold-change calculations, p-values (t-test/ANOVA), and visualization tools (e.g., heatmaps, pathway mapping). Advanced options include PCA plots and metabolic network diagrams to highlight group-specific trends.
Do you support longitudinal studies with repeated sampling?
Yes, we ensure batch-to-batch consistency using standardized protocols and inter-batch calibration. Time-course data are aligned with kinetic models upon request.
What if my sample type is not listed in your validated matrices?
Contact us for a feasibility assessment. We routinely develop methods for novel matrices (e.g., plant root exudates, bioreactor fluids) and provide spike-recovery validation data before full analysis.
Learn about other Q&A.
Malonic Acid Analysis Service Case Study

Title: Extending Metabolomic Studies of Apis mellifera Venom: LC-MS-Based Targeted Analysis of Organic Acids
Journal: Toxins
Published: 2019
- Background
- Methods
- Results
- Reference
Honeybee venom (HBV) contains various biologically active compounds, including peptides, enzymes, and small molecules. While previous research has extensively studied peptides and enzymes, low-molecular-weight organic acids have remained largely unexplored. These organic acids, despite their minor proportion in HBV, may contribute to its pharmacological effects and therapeutic potential. With the advancement of omics technologies, particularly metabolomics using LC-MS/MS, this study aims to provide the first targeted, quantitative analysis of a panel of organic acids in HBV. This contributes to a better understanding of venom composition and supports the development of HBV-based medical applications.
This study utilized a highly sensitive targeted metabolomics approach using HPLC-MS/MS to quantify seven organic acids in honeybee venom (HBV).
1. Reagents & Standards: All analytes and isotope-labeled internal standards were sourced from Sigma-Aldrich.
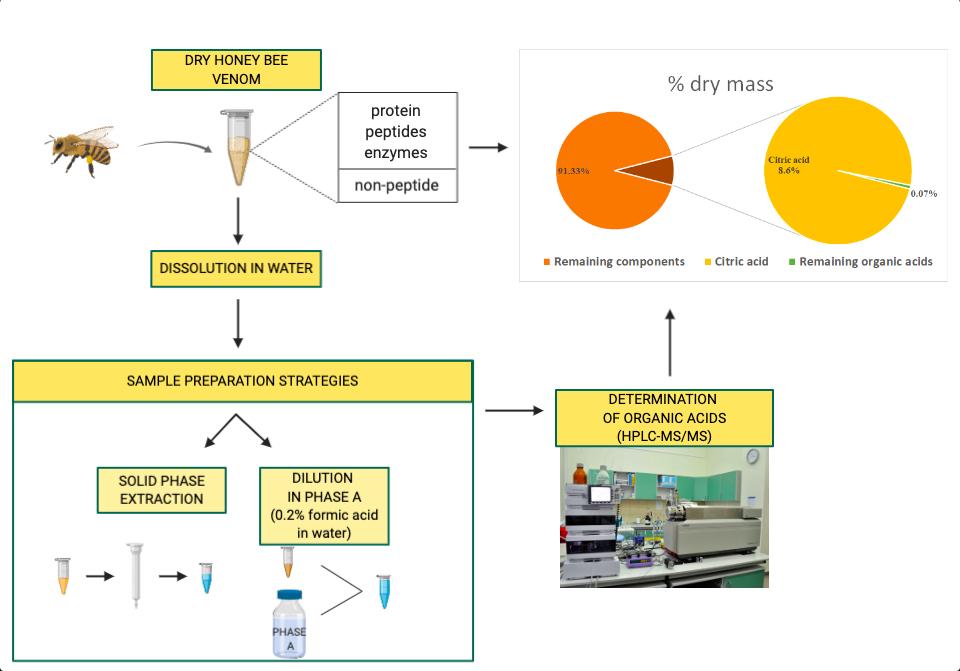
2. Sample Collection: HBV was collected from Apis mellifera via electrical stimulation in 2006, 2017, and 2018. Samples were dried and stored at −80 °C.
3. Sample Preparation:
- Solid Phase Extraction (SPE) was used for analytes at low concentrations (e.g., glutaric acid, kynurenic acid).
- Dilute-and-shoot method was used for high-abundance acids (e.g., citric acid, malic acid).
4. Chromatography: LC separation was performed on a Synergi Hydro-RP column under a formic acid-methanol gradient.
5. Mass Spectrometry: A 4000 QTRAP system operated in negative ESI mode with MRM detection, enabling both quantification and identity confirmation.
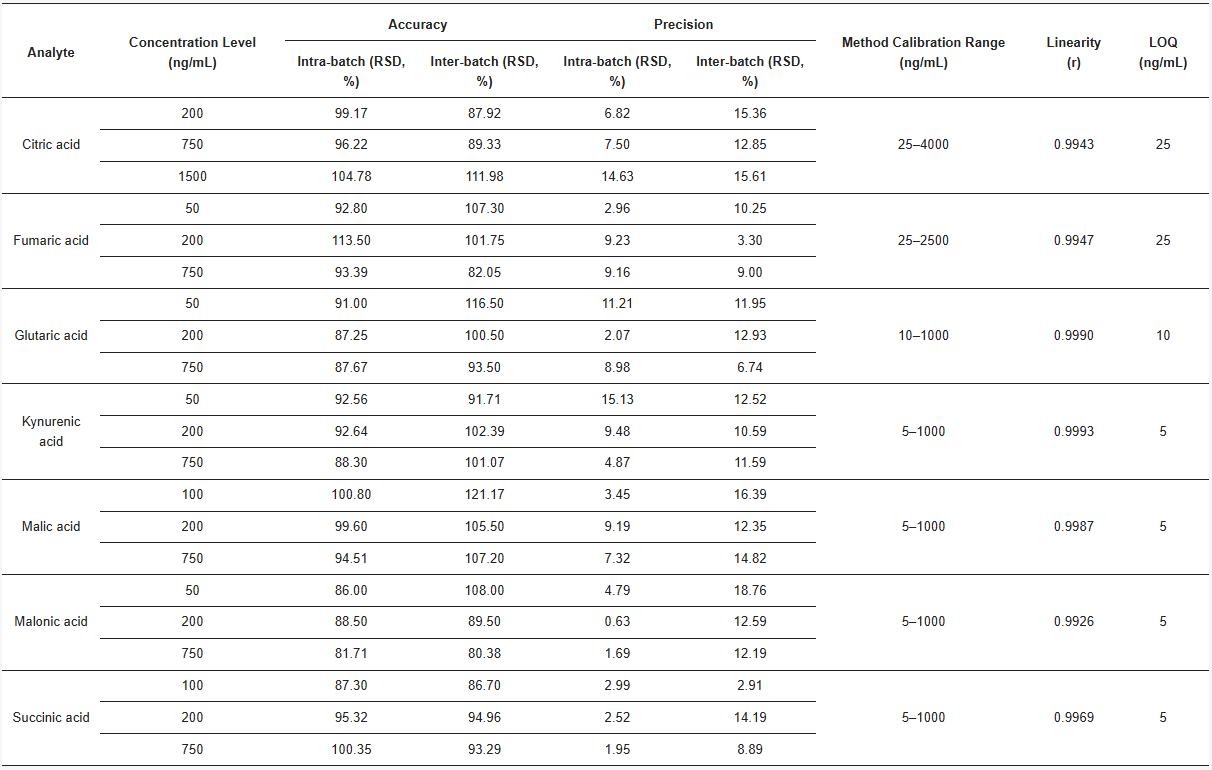
6. Validation: The method was validated for linearity, accuracy, precision, and LOQ, with calibration curves showing r ≥ 0.993. LOQs ranged from 5–25 ng/mL. Intra-/inter-batch precision and accuracy met international validation standards.
 Metabolomic Studies of Apis mellifera Venom
Metabolomic Studies of Apis mellifera Venom
Creative Proteomics Can Support This Research by Providing the Following Malonic Acid-Focused Services:
Based on this study, Creative Proteomics can offer:
- Targeted metabolomics using LC-MS/MS for small molecule profiling in complex matrices like venoms.
- Custom method development and validation for specific metabolites (e.g., organic acids).
- Quantitative analysis with high sensitivity and reproducibility, including low ng/mL detection limits.
- Comparative metabolomic profiling across sample batches or storage conditions.
- Sample preparation optimization, including SPE and dilute-and-shoot protocols.
A targeted HPLC-MS/MS method was successfully developed and validated for quantifying seven organic acids in honeybee venom (HBV): citric, malic, succinic, fumaric, malonic, glutaric, and kynurenic acids. The method demonstrated good linearity (r ≥ 0.993), precision (RSD ≤ 16.39%), and accuracy (80.38%–121.17%). Citric acid was the most abundant, constituting up to 86 mg/g of HBV dry weight and over 99% of total measured organic acids. Succinic acid showed the greatest year-to-year variation (CV = 58.4%), while malonic acid was the most stable (CV = 4.72%). No clear degradation trend was observed in older samples (2006), indicating good long-term stability. Significant intra-year variability suggests biological and environmental influences rather than storage effects.
Table. Validation parameters for HPLC-MS/MS method
Reference
- Pawlak, Magdalena, et al. "Extending metabolomic studies of Apis mellifera venom: LC-MS-based targeted analysis of organic acids." Toxins 12.1 (2019): 14. https://doi.org/10.3390/toxins12010014.
Publications
Here are some publications in Metabolomics research from our clients:

- Thermotolerance capabilities, blood metabolomics, and mammary gland hemodynamics and transcriptomic profiles of slick-haired Holstein cattle during mid lactation in Puerto Rico. 2024. https://doi.org/10.3168/jds.2023-23878
- Enhance trial: effects of NAD3® on hallmarks of aging and clinical endpoints of health in middle aged adults: a subset analysis focused on blood cell NAD+ concentrations and lipid metabolism. 2022. https://doi.org/10.3390/physiologia2010002
- Characterization of a novel AraC/XylS-regulated family of N-acyltransferases in pathogens of the order Enterobacterales. 2020. https://doi.org/10.1371/journal.ppat.1008776
- Disruption of CYCLOPHILIN 38 function reveals a photosynthesis-dependent systemic signal controlling lateral root emergence. 2020. https://doi.org/10.1101/2020.03.11.985820
- Lipin-1 regulates lipid catabolism in pro-resolving macrophages. 2020. https://doi.org/10.1101/2020.06.03.121293






