Kinetin Analysis Service
Creative Proteomics provides high-sensitivity kinetin analysis with detection limits as low as 0.01 ng/mL and >98% reproducibility, powered by advanced LC-MS/MS platforms. Our services support agricultural and pharmaceutical research through precise cytokinin quantification, metabolic pathway mapping, and customized workflows—helping clients optimize tissue culture, enhance crop resilience, and advance metabolic research.
Submit Your Request Now
×- What We Provide
- Advantage
- Workflow
- Technology Platform
- Sample Requirements
- FAQ
- Case Study
What is Kinetin?
Kinetin is a cytokinin-class phytohormone that stimulates cell division in plant roots and shoots, first isolated from autoclaved herring sperm DNA by Skoog, a pioneer in plant growth regulators. Structurally defined as C10H9N5O, it is sparingly soluble in cold water but freely soluble in acidic or alkaline solutions. Initially thought to be synthetic, kinetin was later identified as a natural component in the DNA of diverse organisms, including coconut water. Its primary role lies in plant tissue culture, where it synergizes with auxins to induce callus formation and shoot regeneration.
Why Analyze Kinetin?
Quantifying kinetin levels is critical for:
- Optimizing plant tissue culture protocols (e.g., cocoa zygotic embryo regeneration).
- Studying metabolic pathways linked to seed germination and stress responses (e.g., drought tolerance in hybrid rice).
- Investigating cytokinin dynamics in developmental processes like apical dominance and nutrient mobilization.
Kinetin Analysis Service Offered by Creative Proteomics
- Quantitative Measurement of Kinetin and Its Metabolites: Accurate quantification of kinetin and its metabolic byproducts through state-of-the-art analytical methods.
- Metabolic Pathway Identification: Using advanced mass spectrometry and chromatography techniques to identify and map the metabolic pathways involving kinetin.
- Kinetin Profiling in Different Sample Types: We analyze kinetin content across a range of sample types, including plant tissues, cell cultures, and biological fluids.
- Metabolite Detection: Identification of the metabolites produced from kinetin's biological activity, providing insight into its broader physiological impact.
- Custom Analytical Solutions: Tailored services that meet the specific needs of each client, whether it's for agricultural applications, pharmaceutical development, or environmental studies.
List of Detected Kinetin and Related Metabolites
| Substance | Type | Description | Metabolic Pathway |
|---|---|---|---|
| Kinetin | Cytokinin | The primary compound being analyzed, a plant growth regulator. | Cytokinin biosynthesis pathway, Cell division pathway |
| Kinetin Riboside | Metabolite | A major metabolite of kinetin involved in cytokinin metabolism. | Cytokinin riboside metabolism, Adenine metabolism |
| N6-Furfuryladenine | Metabolite | A derivative of kinetin, involved in stress response and metabolism. | Cytokinin metabolism, Stress response pathways |
| N6-(Δ2-Isopentenyl) Adenine | Metabolite | A compound produced via the isopentenylation of adenine, part of cytokinin biosynthesis. | Isopentenylation pathway, Cytokinin biosynthesis |
| Trans-zeatin | Metabolite | A cytokinin derivative important in regulating plant growth and development. | Cytokinin biosynthesis, Plant growth regulation |
| Cis-zeatin | Metabolite | A form of zeatin, part of cytokinin's biological activity in plants. | Cytokinin biosynthesis, Plant metabolism |
| Isopentenyl Adenine | Metabolite | Related to cytokinin biosynthesis and essential for plant growth regulation. | Cytokinin biosynthesis, Adenine metabolism |
| N6-Benzyladenine | Metabolite | A synthetic cytokinin analog, used in plant tissue culture. | Cytokinin biosynthesis pathway, Plant tissue culture |
| Kinetin Glucoside | Metabolite | A conjugate of kinetin, often involved in plant metabolism and stress response. | Cytokinin conjugation, Metabolism under stress |
| N6-(3-Methylthio) adenine | Metabolite | A metabolite involved in the sulfur-modified pathway of cytokinin biosynthesis. | Sulfur modification in cytokinin metabolism |
Advantages of Kinetin Assay
- Ultra-Low Detection Limit: Detects kinetin and related metabolites at concentrations as low as 0.1 ng/mL using LC-MS/MS.
- High Quantification Accuracy: Quantification accuracy with a relative standard deviation (RSD) of<5% across replicates.
- Broad Dynamic Range: Reliable quantification across 4–5 orders of magnitude, ensuring accurate detection in diverse sample types.
- High Sample Throughput: Capable of processing 96 samples per batch, allowing for efficient large-scale studies.
- Comprehensive Metabolite Coverage: Detects over 10 kinetin-related metabolites within a single run.
- Excellent Reproducibility: Technical reproducibility with >98% consistency between replicate measurements.
- Automated Data Analysis: Integrated software pipelines provide normalized and statistically validated results with<2% data loss.
Workflow for Kinetin Analysis Service
Technology Platform for Kinetin Analysis Service
Sciex QTRAP® 6500+: Offers MRM³ for resolving kinetin-9-glucoside isomers; SelexION+ minimizes matrix effects—ideal for metabolic pathway mapping and stress-response studies.
Agilent 6495C Triple Quad: Fast 20 ms polarity switching and matrix tolerance—perfect for analyzing kinetin in soil and water environments.
Agilent 1290 Infinity II: Supports up to 1,200 bar pressure—efficient for high-throughput analysis of viscous plant extracts in tissue culture research.

SCIEX Triple Quad™ 6500+ (Figure from Sciex)

Agilent 6495C Triple quadrupole (Figure from Agilent)

Agilent 1260 Infinity II HPLC (Fig from Agilent)
Sample Requirements for Kinetin Analysis Service
| Sample Type | Minimum Sample Amount | Sample Preparation Requirements |
|---|---|---|
| Plant Tissue (e.g., leaf, root) | 50 mg | Fresh or freeze-dried tissue is recommended. |
| Cell Culture | 1 mL | Cells should be collected in a buffer solution. |
| Biological Fluids | 500 µL | Serum, plasma, or urine accepted; fresh or frozen. |
| Soil Extract | 10 g | Freshly collected samples should be properly preserved. |
Applications of Kinetin Assay Service
Botanical Quality Control
Authenticate herbal materials and validate standardization levels.
Plant Metabolomics
Understand diterpenoid metabolic pathways in plant systems.
Synthetic Biology Monitoring
Validate cryptotanshinone production in engineered organisms.
Natural Product Research
Identify novel analogs and investigate biosynthetic logic.
Natural Product Research
Identify novel analogs and investigate biosynthetic logic.
Agronomic Phenotyping
Characterize chemotypes across Salvia cultivars and mutants.
FAQ of Kinetin Analysis Service
What measures are taken to ensure consistency in batch-to-batch analysis of kinetin?
We implement rigorous internal quality controls (QCs), including spiked reference samples and blank matrix runs, with inter-batch variability maintained below 5%. Data normalization is performed using MultiQuant™ software to correct for instrumental drift.
Can kinetin be analyzed in samples with high lipid or protein content (e.g., seeds or latex)?
Yes. Our Agilent 6495C Triple Quad system utilizes Jet Stream ionization to minimize matrix interference, coupled with SPE-based lipid/protein removal protocols validated for oilseed and latex samples.
How are kinetin isomers (e.g., cis- vs. trans-kinetin) distinguished in your analysis?
The Sciex QTRAP® 6500+ employs MRM³ scans with ion mobility separation (SelexION+) to resolve isomers, achieving >95% specificity even in root exudate matrices.
Do you provide guidance on kinetin extraction methods for specific plant organs (e.g., roots vs. leaves)?
Absolutely. We offer free protocol customization based on tissue type—e.g., acidified methanol for leaves, or liquid-liquid partitioning for roots—to maximize recovery rates (typically 85–92%).
What file formats are included in the final deliverables?
Results are delivered as a PDF report with raw data in .CSV format (peak areas, retention times) and annotated chromatograms in .CDF or .mzML formats for third-party software compatibility.
Can I track my sample's analysis progress in real time?
Yes. Clients receive a secure login to our LIMS portal for live updates, including QC checkpoints, instrument status, and preliminary data previews.
How do you handle samples suspected of kinetin degradation during shipping?
We perform stability checks upon receipt (e.g., spike recovery tests) and notify clients immediately if degradation is detected. A complimentary re-extraction service is offered for validated storage failures.
Are there discounts for long-term or high-volume projects?
Yes. Volume-based pricing is available for projects exceeding 500 samples/year, with discounts up to 20%. Contact our sales team for a tailored quotation.
What is your policy for retesting ambiguous results?
Ambiguous results (e.g., co-eluting peaks) are retested at no additional cost using orthogonal methods, such as high-resolution Q-TOF MS, to confirm kinetin identity.
Learn about other Q&A.
Kinetin Analysis Service Case Study

Title: Metabolic signatures of germination triggered by kinetin in Medicago truncatula
Journal: Scientific Reports
Published: 2019
- Background
- Materials & Methods
- Results
- Reference
Seed germination involves complex physiological and metabolic transitions, particularly during the shift from a dry, dormant state to an active, hydrated state. This process is challenged by oxidative and genotoxic stress due to reactive oxygen species (ROS) and DNA damage. While the role of phytohormones like gibberellins and abscisic acid in regulating germination is well documented, the function of cytokinins—specifically kinetin—remains less explored. Cytokinins are known to support cell division, stress tolerance, and seed development, but their impact on seed vigor and pre-germinative metabolism is still under investigation. The legume Medicago truncatula, a model species with high relevance for agriculture and breeding, presents an ideal system to study these effects. This study employs non-targeted metabolomics alongside molecular and biochemical assays to uncover how kinetin influences seed germination and related stress responses, aiming to identify metabolic signatures that may contribute to improved seed vigor and resilience.
Plant Materials & Treatments: M. truncatula seeds (Jemalong cultivar) were sown in Petri dishes with distilled water (control) or 0.5 mM kinetin. Seeds were imbibed at 22°C under light conditions with a 16/8h photoperiod and 70-80% humidity. Germination was assessed by radicle protrusion, and the time to 50% germination (T50) was calculated. Fresh and dry weights of seeds and seedlings were measured.
Metabolomic Profiling: Non-targeted profiling was performed using UHPLC/MS/MS and GC/MS platforms on methanol extracts of seed samples. Data were processed by Metabolon Inc. (Durham, NC) and metabolites were identified using a reference library. Statistical analysis included Welch's t-tests for differential metabolite accumulation and PCA for data visualization.
RNA Extraction & qRT-PCR: RNA was extracted from seeds at 2h, 8h, and radicle protrusion stages. cDNA was synthesized, and qRT-PCR was performed using Maxima SYBR Green Master Mix. Relative gene expression was quantified using the Pfaffl method with MtPDF2 as a reference gene.
Comet Assay: Radicles were harvested at the radicle protrusion stage, frozen in liquid N2, and nuclei were extracted. DNA strand breaks were assessed by electrophoresis and DAPI staining. Nucleoids were classified and quantified under a fluorescence microscope.
Electron Paramagnetic Resonance (EPR): EPR analysis was conducted on seeds at 2h, 8h, and radicle protrusion stages. Spectra were recorded at 120K using a Bruker EMX-10/12 spectrometer. Radicals were characterized by intensity measurements and g-values.
Statistical Analysis: Germination data were analyzed using a two-tailed t-test. qRT-PCR and comet assay data were analyzed using Student's t-test. Statistically significant differences were considered at P ≤ 0.05.
- Kinetin (0.5 mM) significantly enhanced seed germination rate and accelerated radicle protrusion and leaf emergence but inhibited root growth.
- Metabolomic analysis revealed that kinetin-induced changes were most evident at the radicle protrusion stage, affecting amino acid, nucleotide, lipid, and carbohydrate metabolism.
- Among 411 identified metabolites, 27 showed significant differential accumulation, mainly reflecting metabolite depletion in kinetin-treated seeds.
- Enrichment analysis indicated major changes in lysine degradation, purine metabolism, phenylalanine/tyrosine, methionine, and branched-chain amino acid degradation.
- Kinetin-treated seeds exhibited increased DNA strand breaks (comet assay), suggesting genotoxic stress.
- ROS levels and radical species remained unchanged (EPR analysis), indicating that oxidative stress was not induced.
- Antioxidant gene expression (e.g., MtAPX, MtMT2, MtTRH1/2) was altered, supporting a stress-related metabolic adjustment.
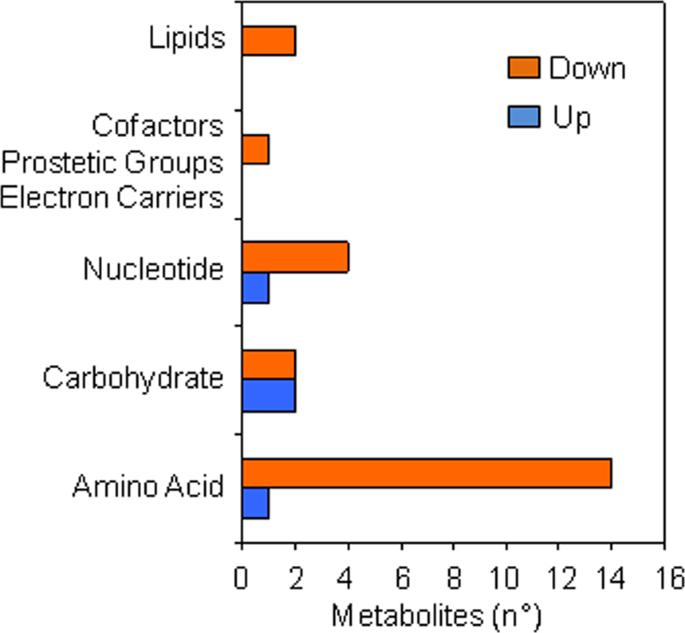 Metabolic super-pathways significantly (P ≤ 0.05 and q ≤ 0.1) changed in response to kinetin treatment at the radicle protrusion stage compared.
Metabolic super-pathways significantly (P ≤ 0.05 and q ≤ 0.1) changed in response to kinetin treatment at the radicle protrusion stage compared.
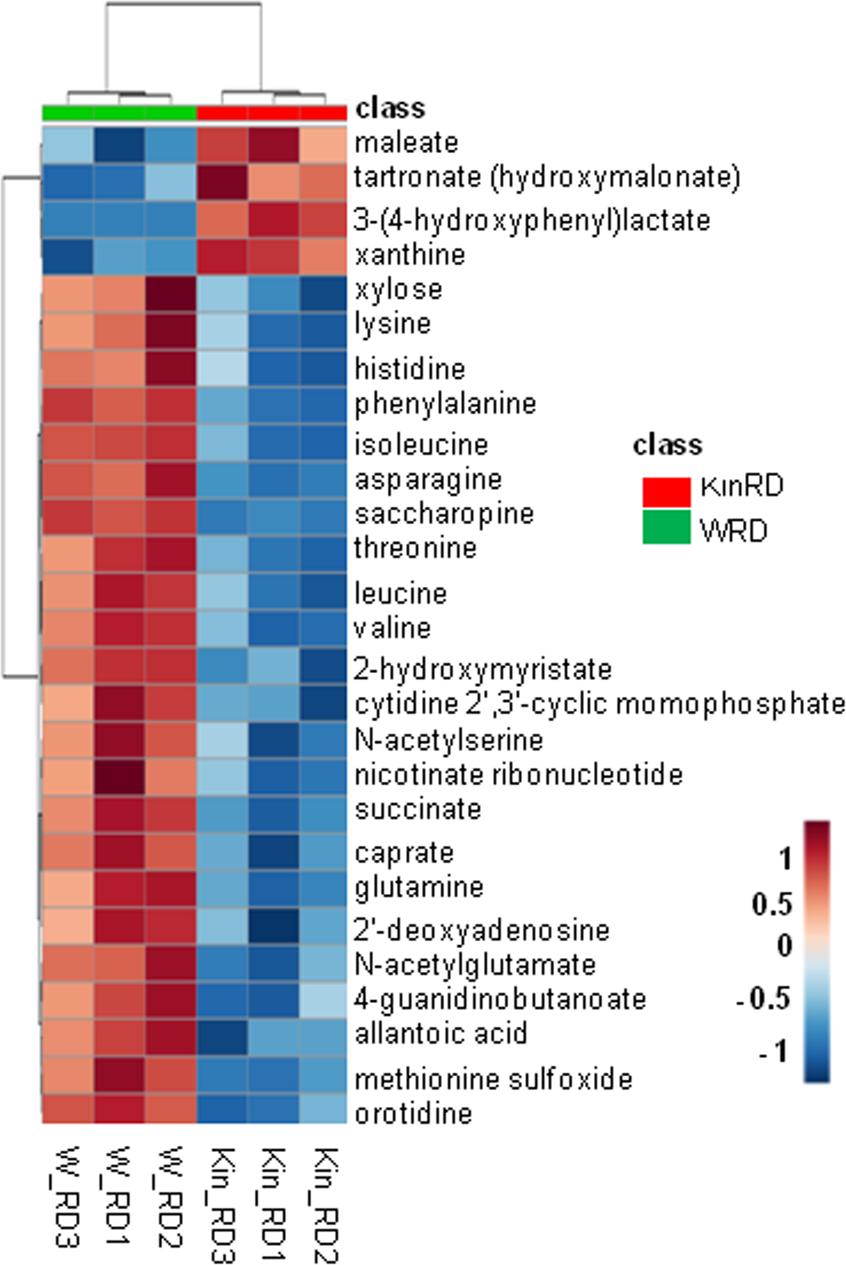 Hierarchical clustering (Euclidean distance measure and Ward clustering algorithm) of 27 metabolites with significant changes (P ≤ 0.05 and q ≤ 0.1) in response to kinetin treatment at radicle protrusion stage (RD).
Hierarchical clustering (Euclidean distance measure and Ward clustering algorithm) of 27 metabolites with significant changes (P ≤ 0.05 and q ≤ 0.1) in response to kinetin treatment at radicle protrusion stage (RD).
Reference
- Araújo, Susana, et al. "Metabolic signatures of germination triggered by kinetin in Medicago truncatula." Scientific Reports 9.1 (2019): 10466. https://doi.org/10.1038/s41598-019-46866-6
Publications
Here are some publications in Metabolomics research from our clients:

- Quantifying forms and functions of intestinal bile acid pools in mice. 2024. https://doi.org/10.1101/2024.02.16.580658
- A comprehensive biochemical characterization of settlement stage leptocephalus larvae of bonefish (Albula vulpes). 2021. https://doi.org/10.1111/jfb.14846
- WI12 Rhg1 interacts with DELLAs and mediates soybean cyst nematode resistance through hormone pathways. 2022. https://doi.org/10.1111/pbi.13709
- Characterization of a novel AraC/XylS-regulated family of N-acyltransferases in pathogens of the order Enterobacterales. 2020. https://doi.org/10.1371/journal.ppat.1008776
- Anti-inflammatory activity of black soldier fly oil associated with modulation of tlr signaling: A metabolomic approach. 2023. https://doi.org/10.3390/ijms241310634






