Isopentenyladenine Analysis Service
Creative Proteomics provides high-precision isopentenyladenine (IPA) analysis services using LC-MS/MS to help researchers uncover plant growth mechanisms, stress responses, and cytokinin metabolism. Our service enables accurate quantification and metabolite profiling across plant, microbial, and environmental samples—supporting agricultural innovation, biotechnology research, and environmental monitoring with minimal sample input and maximum data clarity.
Submit Your Request Now
×- What We Provide
- Advantage
- Workflow
- Technology Platform
- Sample Requirements
- FAQ
- Case Study
- Publications
What is Isopentenyladenine?
Isopentenyladenine (IPA) is a naturally occurring purine base and cytokinin hormone derivative found in plants, fungi, and some bacteria. It plays a critical role in regulating cell division, growth, and response to environmental stress. As a member of the class of compounds known as cytokinins, IPA is essential for maintaining plant health, influencing various physiological processes such as aging, chloroplast development, and the response to abiotic stress. Beyond plants, IPA and its derivatives are implicated in various metabolic pathways in both plants and microorganisms, making its analysis valuable in multiple research fields.
Why Is Isopentenyladenine Analysis Needed?
Monitoring IPA levels provides insights into plant health, metabolic shifts, and environmental adaptations. Analyzing IPA and its metabolites can help researchers investigate pathways related to:
- Plant Growth Regulation: Understanding how IPA affects cytokinin activity and its interplay with other hormones.
- Stress Responses: Investigating how IPA modulates plant responses to abiotic stresses like drought and salinity.
- Biological Pathways: Studying IPA's role in purine metabolism and cytokinin biosynthesis in microorganisms and plants.
- Agriculture & Biotechnology: Optimizing IPA levels for improved crop yield and stress resistance.
Isopentenyladenine Analysis Service Offered by Creative Proteomics
Quantification of Isopentenyladenine
Accurate quantification of IPA concentrations in various biological samples, such as plant tissues, microbial cultures, and environmental samples, using high-sensitivity analytical techniques like LC-MS/MS and HPLC.
Identification of Isopentenyladenine Metabolites
Detection and identification of IPA metabolites, including isopentenyladenosine, zeatin, kinetin, and other related compounds, through advanced LC-MS/MS or HPLC methods, providing a comprehensive analysis of IPA metabolism.
IPA Metabolic Pathway Analysis
Mapping the metabolic pathways involving Isopentenyladenine in plants and microorganisms, leveraging metabolomics data to understand its biological function within cellular networks.
Time-course Analysis of IPA Levels
Monitoring the dynamic changes in IPA levels under various experimental conditions, such as abiotic stress treatments or hormone application, to understand its role in stress responses and growth regulation over time.
Cross-species Comparative IPA Analysis
Comparative analysis of IPA levels across different species (e.g., Arabidopsis, maize, wheat, and various microorganisms), providing insights into interspecies variations in IPA metabolism and adaptation mechanisms.
Plant IPA Response Analysis
Investigating the changes in IPA levels in plants under different environmental stress conditions (e.g., drought, salinity, temperature fluctuations), aiding in research related to plant stress tolerance and adaptation mechanisms.
Environmental Sample IPA Analysis
Analyzing IPA concentrations in environmental samples such as soil, water, and air to assess the impact of environmental factors or agricultural practices on IPA levels in natural ecosystems.
List of Detected Isopentenyladenine and Related Metabolites
| Analyte | Class | Metabolic Pathway |
|---|---|---|
| Isopentenyladenine | Purine Cytokinin | Cytokinin Biosynthesis Pathway |
| Isopentenyladenosine | Cytokinin Derivative | Purine Metabolism Pathway |
| Zeatin | Cytokinin | Cytokinin Biosynthesis Pathway |
| Kinetin | Cytokinin | Cytokinin Biosynthesis Pathway |
| Isopentenyladenine Glucoside | IPA Derivative | Glucosylation in Cytokinin Pathway |
| Adenosine | Purine Nucleoside | Purine Metabolism |
| Trans-zeatin | Cytokinin | Cytokinin Biosynthesis Pathway |
| cis-zeatin | Cytokinin | Cytokinin Biosynthesis Pathway |
| N6-(Δ2-Isopentenyl) adenosine | Cytokinin Derivative | Purine Metabolism |
| Isopentenylthiazole | Cytokinin Metabolite | Cytokinin Biosynthesis/Metabolism |
| Dihydrozeatin | Cytokinin Metabolite | Cytokinin Biosynthesis Pathway |
Advantages of Isopentenyladenine Assay
- Ultra-Low Detection Limits: Detection sensitivity down to 5 pg/mL using LC-MS/MS (Thermo Scientific Q Exactive), enabling accurate quantification of IPA even in trace concentrations.
- High Quantification Accuracy and Precision: Intra-assay coefficient of variation (CV) consistently<5%; inter-assay CV <10%, ensuring reproducibility and consistency across batches and projects.
- Wide Linear Dynamic Range: Quantification range spans 5 pg/mL to 1000 ng/mL, supporting a broad variety of experimental designs from low-concentration signaling studies to high-dose response profiling.
- Multiplex Detection Capability: Simultaneous detection of 12+ IPA-related metabolites in a single run without cross-interference, reducing cost and improving throughput for pathway studies.
- Minimal Sample Requirement: Only 50 mg of tissue or 1 mL of culture needed, minimizing the demand on rare or limited samples while maintaining full analytical fidelity.
- Advanced Chromatographic Separation: Peak resolution of >1.5 between IPA and structurally similar cytokinins using high-efficiency UHPLC columns, ensuring reliable metabolite discrimination.
- Comprehensive Data Output: Each analysis includes raw data (mzXML, CSV), calibration curves (R² > 0.998), quantification tables, and pathway maps, supporting full transparency and downstream bioinformatics.
Workflow for Isopentenyladenine Analysis Service
Technology Platform for Isopentenyladenine Analysis Service
Primary Platform: LC-MS/MS using Sciex QTRAP 6500+ offers femtomole-level sensitivity and excellent reproducibility.
Chromatographic System: UHPLC system (Agilent 1290 Infinity II) equipped with C18 reversed-phase columns for sharp peak resolution.

SCIEX Triple Quad™ 6500+ (Figure from Sciex)

Agilent 1260 Infinity II HPLC (Fig from Agilent)
Sample Requirements for Isopentenyladenine Analysis Service
| Sample Type | Minimum Amount Required | Recommended Amount | Preservation Method |
|---|---|---|---|
| Plant Tissue (Fresh/Frozen) | 50 mg | 200 mg | Freeze in liquid nitrogen, store at –80°C |
| Microbial Culture (Liquid) | 1 mL | 5 mL | Centrifuge and freeze pellet at –80°C |
| Environmental Water Sample | 10 mL | 50 mL | Filter if possible, store at –20°C |
| Soil Extract | 100 mg | 500 mg | Dry and store in sealed container at –20°C |
| Serum / Plasma | 200 µL | 500 µL | Store at –80°C |
| Seed Extract | 50 mg | 200 mg | Dry and store at –20°C |
FAQ of Isopentenyladenine Analysis Service
Can you analyze samples from non-model plants or exotic species?
Yes. Our methods are validated for a wide range of plant species, including non-model and rare plants. We recommend submitting a small pilot batch if the species is highly unique or chemically complex.
What is the best way to ship samples for IPA analysis?
For biological samples such as plant tissue or microbial pellets, ship on dry ice using overnight courier. Environmental samples can be shipped on ice packs if frozen storage is not feasible. Avoid repeated freeze-thaw cycles.
Can I submit samples in an extract form rather than raw material?
Yes. We accept both raw biological material and pre-processed extracts. Please provide detailed information about your extraction protocol to ensure compatibility with our detection systems.
Do you offer method customization based on specific research needs?
Absolutely. We can adjust analytical parameters such as detection range, metabolite panels, and data output formats depending on your experimental design. Contact us to discuss custom configurations.
Is biological replication necessary for IPA analysis?
Yes. For statistically meaningful data, we recommend submitting at least three biological replicates per condition. Technical replicates are optional but can improve confidence in low-abundance measurements.
Can you help with data interpretation and pathway mapping?
Yes. In addition to raw data and quantification tables, we offer optional data interpretation services, including pathway mapping, statistical comparison, and correlation analysis with other omics datasets.
What should I do if my sample volume is below the recommended minimum?
Contact us before submitting. In many cases, we can adjust extraction protocols to accommodate low-input samples, although detection limits and reproducibility may be impacted.
Do you store the leftover samples after analysis?
Yes. Residual samples are stored at –80°C for up to 30 days post-analysis. If you wish to have them returned or stored for longer periods, please indicate this on your submission form.
Can I combine IPA analysis with other hormone or metabolite profiling?
Yes. We offer multiplex analysis packages that include cytokinins, auxins, gibberellins, and ABA. This is particularly useful for systems biology or stress-response studies.
What is the file format of the data I will receive?
You will receive a comprehensive report including raw data files (e.g., mzXML, CSV), processed quantification tables, calibration curves, and optional annotated pathway diagrams in PDF format.
Learn about other Q&A.
Isopentenyladenine Analysis Service Case Study

Title: Tandem mass spectrometry identification and LC–MS quantification of intact cytokinin nucleotides in K-562 human leukemia cells
Journal: Analytical and Bioanalytical Chemistry
Published: 2010
- Background
- Materials & Methods
- Results
Cytokinins are a group of purine-based plant hormones that regulate cell division and differentiation. Although extensively studied in plants, their roles and biosynthesis pathways have not been fully elucidated at the nucleotide level due to analytical limitations. Recent findings suggest that cytokinin ribosides and their phosphorylated forms may also exhibit cytotoxicity in human cancer cells. This study presents a novel HPLC–MS method for the direct detection and quantification of intact cytokinin nucleotides (mono-, di-, and triphosphates) in K-562 human leukemia cells. The method offers improved sensitivity, retention, and analytical performance without requiring nucleotide degradation or derivatization, filling a critical gap in cytokinin biosynthesis and function analysis.
Chemicals:
Cytokinin standards were purchased or synthesized as described. Analytical-grade solvents and reagents were used.
Instrumentation:
HPLC–MS and MS/MS analyses were performed using Waters and Bruker systems with ESI detection. NMR spectra were recorded on a 300 MHz Bruker spectrometer.
Standard Synthesis:
Monophosphates were prepared via nucleophilic substitution of chloropurine riboside monophosphates. Diphosphates/triphosphates were synthesized under alkaline conditions and purified by HPLC.
HPLC–MS Analysis:
Compounds were separated on C18 columns using methanol–ammonium salt gradients and detected via PDA and ESI–MS in negative SIM mode.
Cell Culture & Treatment:
K-562 cells were cultured in DMEM and treated with 10 μM iPR for 12 h.
Sample Preparation:
Cells were extracted with TCA, deproteinated, back-extracted with ether, lyophilized, and analyzed by LC–MS/MS.
Metabolite Identification & Quantification:
Metabolites were identified by LC–MS/MS and quantified using isotope dilution LC–MS. Method validation included LOD, linearity, recovery, and precision.
Stability Test:
Analyte stability was assessed at different pH and temperatures over 24 h by LC–MS.
Nine cytokinin nucleotides, including mono-, di-, and triphosphates, were synthesized and verified by ESI–MS, UV, and NMR. Using a high-pH-stable hybrid column and ammonium acetate buffer (20 mM, pH 10.0), baseline separation of cis–trans isomers and nucleotides with varying phosphate groups was achieved. Trans-zeatin phosphates consistently eluted before their cis counterparts, and iP nucleotides showed the longest retention.
Method optimization favored high-pH over ion-pairing chromatography due to greater robustness and column lifespan (>500 injections without degradation). Negative ionization mode was optimal, with clear fragmentation patterns observed for iPMP, iPDP, and iPTP.
Triphosphates and diphosphates showed lower recovery (70–80%) compared to monophosphates (97%), attributed to lower stability, though this did not compromise quantification accuracy due to isotope-labeled internal standards. Stability tests confirmed analyte integrity over 24 hours at relevant conditions.
The method exhibited high sensitivity (femtomole LODs) and linearity (1–1000 pmol) across all nine compounds. In iPR-treated K-562 cells, iPMP, iPDP, and iPTP were detected, with iPDP and iPTP representing less than 1% of total iP-derived phosphates.
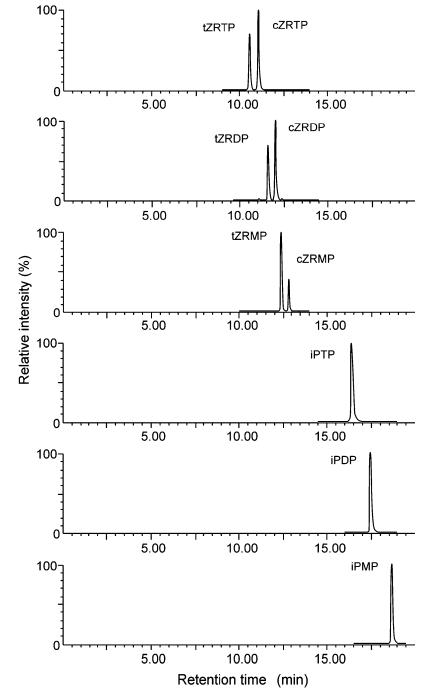 Reconstructed SIM chromatograms of CK nucleotides under optimal chromatographic conditions.
Reconstructed SIM chromatograms of CK nucleotides under optimal chromatographic conditions.
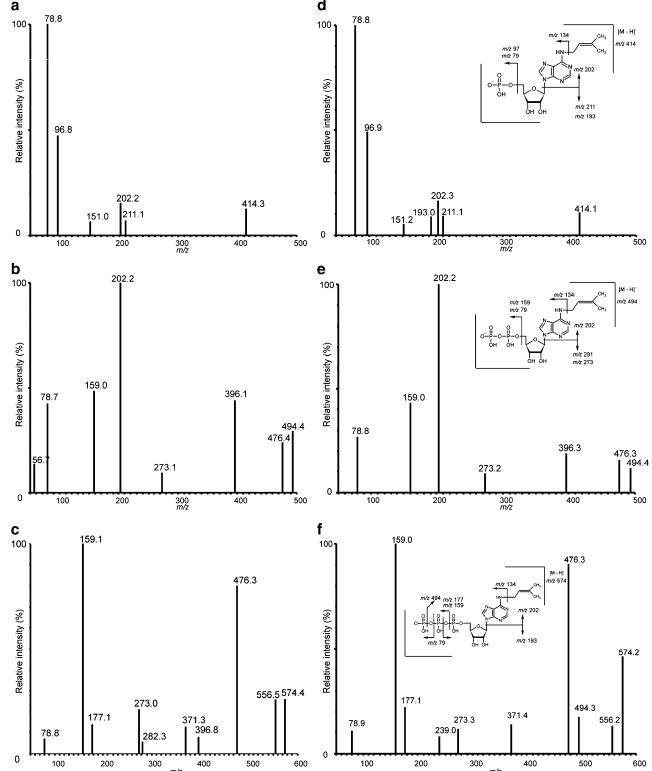 MS/MS spectra and fragmentation patterns obtained for the putative intracellular metabolites of iPR, a iPMP, b iPDP, and c iPTP, extracted from treated cells and the standard solutions of iPMP (d), iPDP (e), and iPTP (f)
MS/MS spectra and fragmentation patterns obtained for the putative intracellular metabolites of iPR, a iPMP, b iPDP, and c iPTP, extracted from treated cells and the standard solutions of iPMP (d), iPDP (e), and iPTP (f)
Reference
- Béres, Tibor, et al. "Tandem mass spectrometry identification and LC–MS quantification of intact cytokinin nucleotides in K-562 human leukemia cells." Analytical and bioanalytical chemistry 398 (2010): 2071-2080. https://doi.org/10.1007/s00216-010-4126-5
Publications
Here are some publications in Metabolomics research from our clients:

- UDP-Glucose/P2Y14 Receptor Signaling Exacerbates Neuronal Apoptosis After Subarachnoid Hemorrhage in Rats. 2024. https://doi.org/10.1161/STROKEAHA.123.044422
- Living in extreme environments: a photosynthetic and desiccation stress tolerance trade-off story, but not for everyone. 2023. https://doi.org/10.22541/au.168311184.42382633/v2
- Untargeted metabolomics reveal sex-specific and non-specific redox-modulating metabolites in kidneys following binge drinking. 2023. https://doi.org/10.1530/REM-23-0005
- Inflammation primes the kidney for recovery by activating AZIN1 A-to-I editing. 2023. https://doi.org/10.1101/2023.11.09.566426
- Exogenous lipase administration alters gut microbiota composition and ameliorates Alzheimer’s disease-like pathology in APP/PS1 mice. 2022. https://doi.org/10.1038/s41598-022-08840-7






