Isoferulic Acid Analysis Service
Creative Proteomics provides high-precision isoferulic acid analysis using advanced UHPLC-MS/MS platforms. We offer targeted quantification, pathway mapping, and isotope tracing to support pharmaceutical research, functional food development, and plant metabolomics. With ultra-sensitive detection (LOD 0.2 ng/mL), CV<5%, and high-throughput capacity (192 samples/day), we deliver fast, accurate, and scalable solutions for complex biological and plant samples.
Submit Your Request Now
×- What We Provide
- Advantage
- Workflow
- Technology Platform
- Sample Requirements
- FAQ
- Case Study
Overview of Isoferulic Acid
Isoferulic acid (3-hydroxy-4-methoxycinnamic acid) is a naturally occurring phenolic compound, predominantly found in plant-based foods and medicinal herbs. Structurally related to ferulic acid, isoferulic acid exhibits potent antioxidant, anti-inflammatory, and anti-glycation activities. Its biological significance stems from its role as a secondary metabolite in various plant metabolic pathways, including the phenylpropanoid and flavonoid biosynthesis pathways.
In plant systems, isoferulic acid often functions as a signaling molecule and a precursor for complex polyphenolic structures. Accurate quantification and profiling of isoferulic acid and its related metabolites are essential in fields such as plant metabolomics, functional food development, and phytochemical research.
Why Analyze Isoferulic Acid?
The need for isoferulic acid analysis arises from its central role in numerous biological processes and industrial applications:
- Quantitative Phytochemical Profiling: Supporting ingredient standardization and quality control in botanicals and food products.
- Metabolic Pathway Elucidation: Deciphering the biosynthetic pathways involving hydroxycinnamic acids.
- Functional Food Development: Establishing correlations between isoferulic acid content and bioactive functionality.
- Biological Response Assessment: Monitoring isoferulic acid levels under different environmental or physiological conditions.
Isoferulic Acid Analysis Service Offered by Creative Proteomics
- Targeted Quantification of Isoferulic Acid: Accurate quantification in complex matrices using UHPLC-MS/MS with authenticated standards.
- Profiling of Isoferulic Acid Derivatives: Detection and relative quantification of structurally related compounds and metabolites.
- Untargeted Phenolic Metabolomics: Comprehensive screening of phenolic acids, including isoferulic acid, for global metabolic insights.
- Metabolic Pathway Annotation: Integration of detected metabolites into KEGG and MetaCyc pathways for functional analysis.
- Stable Isotope Tracing (¹³C/²H): Analysis of isotope-labeled isoferulic acid to study metabolic flux and biosynthetic routes.
- Custom Method Development: Tailored extraction and detection protocols for specific sample types or experimental needs.
- Statistical Analysis and Data Interpretation: Group comparison, significance testing, and multivariate analysis with professional visualization.
Detected Isoferulic Acid and Related Analytes
| Compound/Metabolite | Chemical Class | Associated Metabolic Pathway |
|---|---|---|
| Isoferulic acid | Hydroxycinnamic acid | Phenylpropanoid biosynthesis |
| Ferulic acid | Hydroxycinnamic acid | Lignin biosynthesis, Phenylpropanoid pathway |
| Caffeic acid | Phenolic acid | Flavonoid biosynthesis, Phenylpropanoid |
| p-Coumaric acid | Phenolic acid | Phenylalanine metabolism |
| Sinapic acid | Methoxylated phenolic | Secondary metabolite biosynthesis |
| Vanillic acid | Aromatic carboxylic acid | Tyrosine and lignin metabolism |
| Coniferyl alcohol | Monolignol | Lignin biosynthesis |
| 3,4-Dihydroxyphenylpropionic acid | Phenylpropanoid derivative | Microbial degradation of aromatic compounds |
| Syringic acid | Methoxylated phenolic | Methoxylation pathway of phenolic acids |
| Chlorogenic acid | Cinnamate ester | Phenylpropanoid metabolism, antioxidant pathway |
| Isoferulic acid glucuronide | Conjugated metabolite | Phase II metabolism (glucuronidation) |
| Isoferulic acid sulfate | Sulfated conjugate | Phase II metabolism (sulfation) |
Advantages of Isoferulic Acid Assay
- Low Detection Limit: Down to 0.2 ng/mL for isoferulic acid using optimized UHPLC-MS/MS method.
- High Quantitative Accuracy: CV < 5% across biological replicates.
- Wide Dynamic Range: Quantification linearity from 0.5 ng/mL to 1000 ng/mL.
- High Throughput: Up to 192 samples processed per day.
- Batch Consistency: Batch-to-batch variance < 3%, ensuring reliable comparisons across experiments.
Workflow for Isoferulic Acid Analysis Service
Technology Platform for Isoferulic Acid Analysis Service
Agilent 6495C Triple Quadrupole LC/MS: Ultra-sensitive quantification of isoferulic acid and metabolites at sub-ng/mL levels, ideal for complex matrices.
Agilent 7890B GC + 5977A MSD: Detection of isoferulic acid derivatives (e.g., TMS) after chemical derivatization, suitable for volatile organic acids in plant/food samples.
Agilent 1260 Infinity II HPLC: Efficient separation and qualitative detection of isoferulic acid, ideal for high-concentration samples and quick screening.

Agilent 6495C Triple Quadrupole (Figure from Agilent)

Agilent 7890B-5977A (Figure from Agilent)

Agilent 1260 Infinity II HPLC (Fig from Agilent)
Sample Requirements for Isoferulic Acid Analysis Service
| Sample Type | Minimum Amount Required | Storage Condition | Notes |
|---|---|---|---|
| Plant tissue (leaves, roots, seeds) | ≥ 100 mg (fresh/freeze-dried) | -80°C | Avoid contamination with soil or resin |
| Food samples (grains, fruits, processed food) | ≥ 200 mg | -80°C | Homogenized, free of additives |
| Biological fluids (plasma, serum, urine) | ≥ 500 µL | -80°C | Collected in anticoagulant-free tubes |
| Fermentation broth | ≥ 500 µL | -80°C | Centrifuge and collect supernatant if needed |
| Cell pellets (microbial/plant cells) | ≥ 1 × 10⁸ cells | -80°C | Washed with PBS before freezing |
| Tissue extracts or metabolites | ≥ 200 µL or equivalent | -80°C | Must indicate extraction solvent |
FAQ of Isoferulic Acid Analysis Service
What is the minimum sample quantity required for analysis?
Typically, 10–50 mg of plant tissue or 1–5 mL of liquid samples (e.g., juice, wine). We optimize protocols for low-yield samples.
How do you handle anthocyanin degradation during shipping?
Ship samples on dry ice or in stabilizing buffers (provided upon request). Avoid light exposure using amber vials.
Can your analysis distinguish between anthocyanin degradation products and native compounds?
Yes, our LC-MS/MS methods separate and quantify degradation markers (e.g., protocatechuic acid) from intact anthocyanins.
Do you support data interpretation for agricultural studies?
We provide statistical correlations between anthocyanin profiles and environmental/growth conditions, including heatmaps and pathway activity scores.
How do I choose between targeted quantification and untargeted profiling?
Targeted is ideal for validating known anthocyanins (e.g., quality control). Untargeted suits novel compound discovery or pathway studies.
Can I combine anthocyanin analysis with other polyphenol quantification?
Yes, our metabolomics platform supports simultaneous profiling of anthocyanins, flavonols, catechins, and phenolic acids.
What software tools do you provide for data visualization?
Interactive PCA plots, volcano plots, and pathway maps via MetaboAnalyst 5.0. Raw data formats (mzML, CSV) are also available.
Do you analyze anthocyanins in processed foods with additives?
Yes, but preservatives like sulfites may interfere. Pre-treatment protocols can minimize matrix effects—contact us for details.
How are results delivered?
Secure online portal access with downloadable PDF reports, Excel files, and annotated chromatograms/spectra.
Can your service identify anthocyanin-binding proteins or complexes?
No—this requires separate proteomics workflows. We focus on free and conjugated anthocyanins in extracts.
Learn about other Q&A.
Isoferulic Acid Analysis Service Case Study

Title: Cimicifugae Rhizoma Extract Attenuates Oxidative Stress and Airway Inflammation via the Upregulation of Nrf2/HO-1/NQO1 and Downregulation of NF-κB Phosphorylation in Ovalbumin-Induced Asthma
Journal: Antioxidants
Published: 2021
- Background
- Methods
- Results
- Conclusion
- Reference
Allergic asthma is a chronic inflammatory disease marked by airway hyperresponsiveness, eosinophilic infiltration, and mucus overproduction, driven by oxidative stress and dysregulated immune responses. The imbalance between reactive oxygen species (ROS) and antioxidant defenses, including downregulation of Nrf2/HO-1/NQO1 pathways and overactivation of NF-κB signaling, contributes significantly to disease progression. Although current treatments exist, they often lack efficacy or have adverse effects.
Cimicifugae Rhizoma, a traditional East Asian medicinal herb known for its antioxidant and anti-inflammatory properties, has been shown to alleviate inflammatory conditions but has not been thoroughly evaluated in the context of allergic asthma. This study investigates the therapeutic potential of Cimicifugae Rhizoma extract (CRE) in a murine model of ovalbumin-induced asthma, focusing on its regulatory effects on oxidative stress signaling pathways.
- Plant Material & Extraction: Cimicifugae Rhizoma was extracted with 70% ethanol using heat-reflux, filtered, concentrated, and freeze-dried.
- Chemical Standards & Reagents: Included isoferulic acid, caffeic acid, ferulic acid, and others, with ≥99% purity.
- HPLC Analysis: Quantitative profiling of CRE using HPLC-DAD, with detection at 320 nm and gradient elution, validated by linear regression, LOD, and LOQ assessment.
- Animal Model: BALB/c mice were sensitized with OVA and treated with CRE (30 or 100 mg/kg) or dexamethasone. AHR was measured using plethysmography.
- BALF & Serum Analysis: Inflammatory cytokines (IL-4, IL-5, IL-13) and OVA-specific IgE were quantified using ELISA.
- Histological & Immunoassays: Lung sections were stained (H&E, PAS), and HO-1/MMP-9 expression assessed by immunohistochemistry and immunofluorescence.
- Western Blot & Zymography: Protein expression of Nrf2, HO-1, NQO1, NF-κB, and MMP-9 evaluated using western blotting and gelatin zymography.
- Statistical Analysis: ANOVA with Dunnett's test; significance at p < 0.05.
Creative Proteomics Can Support This Research by Providing the Following Isoferulic Acid-Focused Services:
- Isoferulic Acid Quantification: Accurate identification and quantification of isoferulic acid and structurally related phenolic acids using advanced HPLC-MS/MS platforms, ensuring high sensitivity and specificity.
- Phytochemical Profiling in Herbal Extracts: Comprehensive detection of bioactive constituents in Cimicifugae Rhizoma, including isoferulic acid, caffeic acid, and ferulic acid, based on validated plant metabolomics workflows.
- Standard Curve Generation & Method Validation: Provision of reference standard calibration, LOD/LOQ determination, and reproducibility assessment, fully aligned with pharmaceutical and herbal compound quantification standards.
- Matrix Optimization for Complex Samples: Customized sample preparation strategies for plant matrices and biological fluids to improve extraction efficiency and signal accuracy for isoferulic acid detection.
- Batch Analysis of Experimental Groups: Quantitative comparison of isoferulic acid content across treated groups (e.g., CRE30 vs. CRE100), facilitating dose-effect correlation analysis.
HPLC analysis of CRE identified five key compounds—caffeic acid, ferulic acid, isoferulic acid, cimicifugic acid B, and F—with isoferulic acid (0.81%) and cimicifugic acid F (1.09%) being the most abundant. In an OVA-induced asthma model, CRE significantly reduced airway hyperresponsiveness (AHR), eosinophil infiltration, mucus production, and OVA-specific IgE levels. CRE also suppressed inflammatory cytokines (IL-4, IL-5, IL-13) and upregulated antioxidant markers (Nrf2, HO-1, NQO1). Furthermore, CRE inhibited NF-κB phosphorylation and MMP-9 expression, both critical mediators of airway inflammation and remodeling.
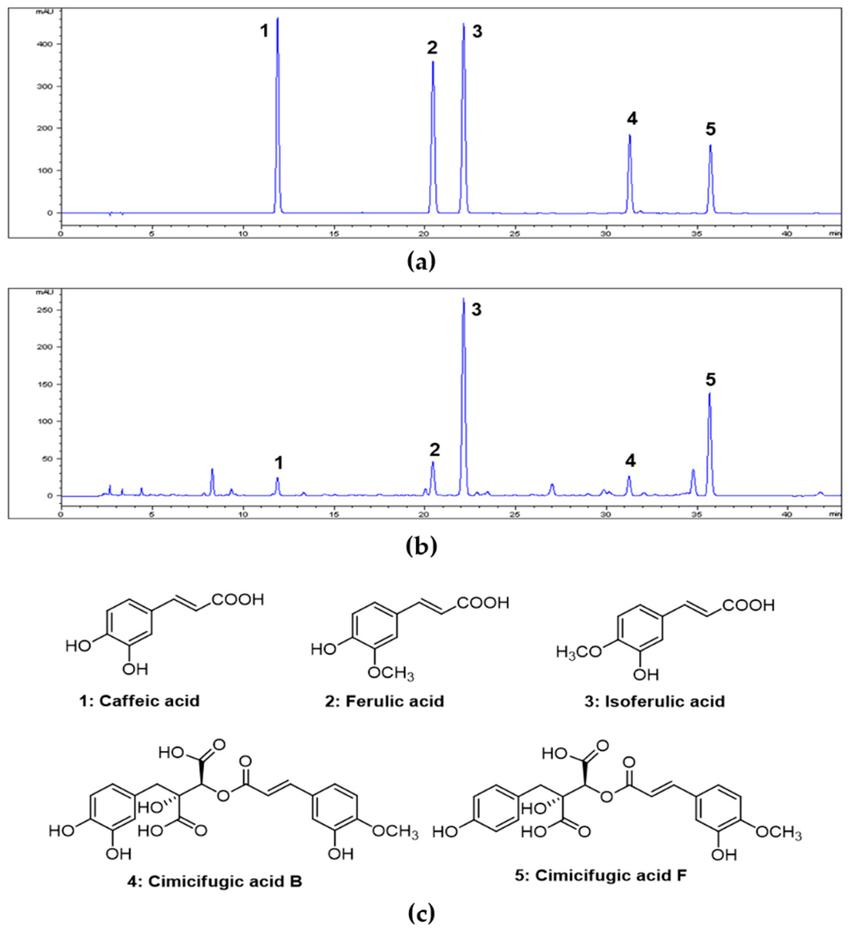 High-performance liquid chromatography (HPLC) chromatograms of five standard compound mixtures (a) and 70% ethanol extract of Cimicifugae Rhizoma (b). Chemical structures of five compounds (c).
High-performance liquid chromatography (HPLC) chromatograms of five standard compound mixtures (a) and 70% ethanol extract of Cimicifugae Rhizoma (b). Chemical structures of five compounds (c).
CRE alleviates asthma symptoms by modulating immune responses and enhancing antioxidant defense. Its effects are associated with the activation of Nrf2/HO-1/NQO1 signaling and suppression of NF-κB/MMP-9-mediated inflammation. These therapeutic actions are likely driven by hydroxycinnamic acid derivatives in CRE, particularly isoferulic, caffeic, and ferulic acids. CRE shows promise as a natural anti-asthmatic agent.
Reference
- Lim, Je-Oh, et al. "Cimicifugae rhizoma extract attenuates oxidative stress and airway inflammation via the upregulation of Nrf2/HO-1/NQO1 and downregulation of NF-κB phosphorylation in ovalbumin-induced asthma." Antioxidants 10.10 (2021): 1626. https://doi.org/10.3390/antiox10101626
Publications
Here are some publications in Metabolomics research from our clients:

- Pan-lysyl oxidase inhibition disrupts fibroinflammatory tumor stroma, rendering cholangiocarcinoma susceptible to chemotherapy. 2024. https://doi.org/10.1097/HC9.0000000000000502
- Glycine supplementation can partially restore oxidative stress-associated glutathione deficiency in ageing cats. 2024. https://doi.org/10.1017/S0007114524000370
- Resting natural killer cell homeostasis relies on tryptophan/NAD+ metabolism and HIF‐1α. 2023. https://doi.org/10.15252/embr.202256156
- Exogenous lipase administration alters gut microbiota composition and ameliorates Alzheimer’s disease-like pathology in APP/PS1 mice. 2022. https://doi.org/10.1038/s41598-022-08840-7
- Thermotolerance capabilities, blood metabolomics, and mammary gland hemodynamics and transcriptomic profiles of slick-haired Holstein cattle during mid lactation in Puerto Rico. 2024. https://doi.org/10.3168/jds.2023-23878







