Omega-6 Fatty Acids Analysis Service
Creative Proteomics offers high-sensitivity omega-6 fatty acids analysis using GC-MS and LC-MS/MS to support lipidomics, product development, and metabolic research.
We help you:
- Quantify over 40 omega-6 fatty acids and bioactive metabolites
- Evaluate omega-6/omega-3 balance in complex biological or nutritional systems
- Monitor lipid profile changes in food, feed, fermentation, or tissue studies
Submit Your Request Now
×
What You Will Receive
- Full quantitative report (ng/mL or µg/g)
- GC-MS & LC-MS/MS chromatograms
- Annotated metabolite list
- Optional PCA / ratio interpretation plots
- Exportable Excel & PDF formats
- What We Provide
- Advantages
- Technology Platform
- Sample Requirements
- Demo
- FAQs
- Case Study
Overview of Omega-6 Fatty Acids
Omega-6 fatty acids are a key class of polyunsaturated fatty acids (PUFAs) that are not synthesized endogenously and must be obtained from dietary sources. As structural components of phospholipid membranes and precursors to potent lipid mediators—including prostaglandins and leukotrienes—omega-6 fatty acids play integral roles in inflammation, cell signaling, and metabolic regulation.
The biological significance of omega-6 lipids lies in their dual capacity: when balanced, they support tissue repair and immune defense; when dysregulated, they contribute to chronic inflammatory states. Understanding omega-6 fatty acid profiles—including both parent compounds such as linoleic acid (LA) and downstream metabolites like arachidonic acid (AA)—is essential for deciphering lipid metabolism, evaluating therapeutic interventions, and ensuring lipid quality in clinical and industrial settings.
Why Analyze Omega-6 Fatty Acids?
Accurate quantification of omega-6 fatty acids and their metabolites supports research and product development across a wide range of scientific fields. Measuring omega-6 profiles enables:
- Investigation of lipid metabolism and nutrient utilization in cells, tissues, and formulated products
- Characterization of fatty acid balance, such as omega-6/omega-3 ratios, to assess the nutritional or biochemical status of a system
- Tracking changes in lipid composition during formulation optimization, fermentation processes, or environmental exposure studies
- Monitoring the generation of omega-6-derived oxylipins and eicosanoids, which are key signaling molecules in stress and immune pathways
- Supporting fatty acid enrichment studies, functional ingredient evaluation, or feedstock analysis in agricultural and industrial contexts
Omega-6 Fatty Acids Analysis Service Offered by Creative Proteomics
- Targeted Quantification via LC-MS/MS: Absolute quantification of NAA using stable isotope-labeled internal standards. Detection limit: < 1 ng/g (tissue).
- Auxin Metabolite Profiling: Simultaneous detection of NAA with natural auxins (e.g., IAA, PAA) and their oxidized or conjugated forms.
- Environmental Residue Testing: Detection of trace NAA in soil, hydroponic solution, and runoff water with validated sample preparation protocols.
- Stability & Degradation Studies: Monitoring NAA metabolism under different light, temperature, and enzymatic conditions in planta.
- Matrix Types Supported: Leaf, root, stem, seed, callus tissue, culture media, soil, and aqueous environmental samples.
Detected Omega-6 Fatty Acids and Related Analytes
| Compound Name | Abbreviation | Class | Related Pathway |
|---|---|---|---|
| 1-Naphthylacetic acid | NAA | Synthetic auxin | Auxin response modulation |
| Indole-3-acetic acid | IAA | Natural auxin | Tryptophan-dependent auxin biosynthesis |
| Indole-3-butyric acid | IBA | Natural auxin | Auxin conjugate and transport pathway |
| Phenylacetic acid | PAA | Natural auxin | Secondary auxin pathway |
| 1-Naphthol | NOL | NAA degradation product | Oxidative auxin breakdown |
| 1-Naphthylacetamide | NAM | Precursor/intermediate | Synthetic auxin metabolism |
| 2-Naphthylacetic acid | 2-NAA | Isomer (inactive) | Structural analog surveillance |
| IAA-Aspartate | IAA-Asp | Conjugated auxin | Auxin inactivation & storage pathway |
| IAA-Glucose | IAA-Glc | Conjugated auxin | Glycosylation and reversible storage |
| OxIAA (Oxindole-3-acetic acid) | OxIAA | Oxidized auxin | Auxin catabolic pathway |
Advantages of Omega-6 Fatty Acids Assay
- High Sensitivity: Detection limits down to 0.05 ng/mL for key oxylipins using LC-MS/MS in negative ion mode.
- Quantitative Reproducibility: CVs < 10% across technical and biological replicates.
- Wide Linear Range: 0.1 ng/mL to 10 µg/mL for fatty acids and metabolites, covering physiological and pathological levels.
- Multiplex Capability: Simultaneous quantification of >40 omega-6 fatty acids and derivatives in a single run.
- Customizable Workflows: Tailored protocols for complex matrices including oils, cell cultures, tissue homogenates, and bioproducts.
- Bioinformatics Support: Optional pathway analysis, fatty acid ratio interpretation, and PCA/PLS-DA clustering for lipidomic datasets.
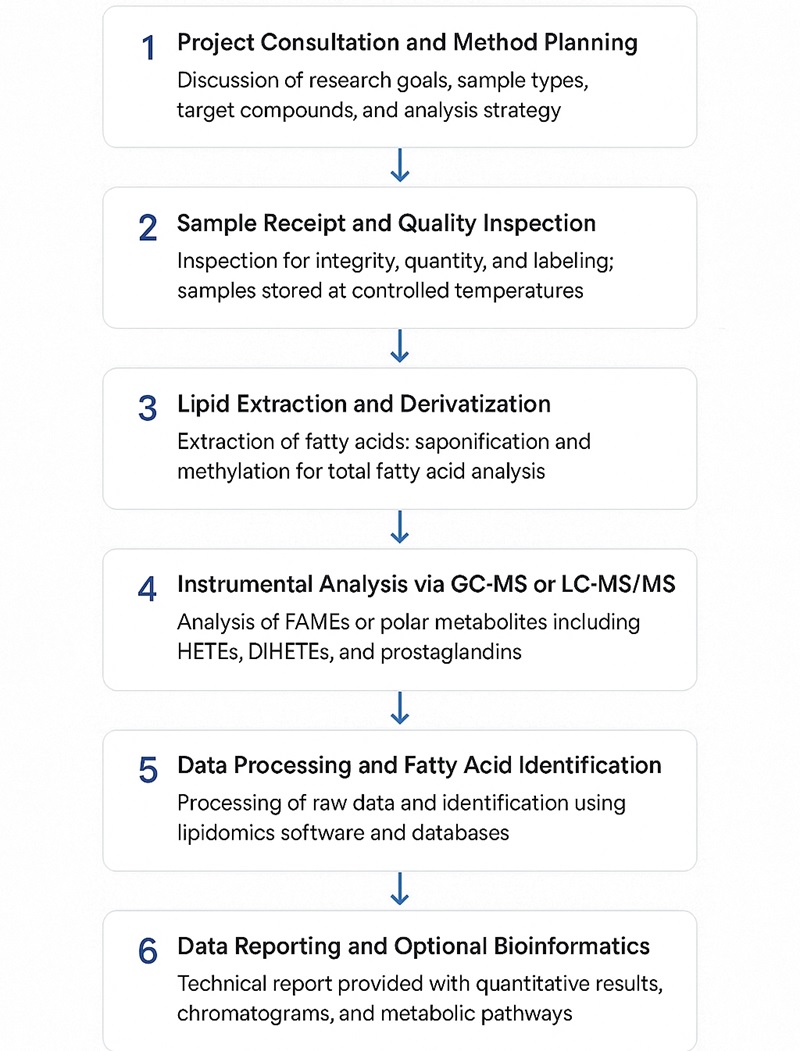
Workflow for Omega-6 Fatty Acids Analysis Service
Technology Platform for Omega-6 Fatty Acids Analysis Service
Agilent 7890B GC coupled with 5977A MSD
Used for the analysis of free and total omega-6 fatty acids in the form of fatty acid methyl esters (FAMEs), enabling precise quantification of LA, AA, DGLA, GLA, and other parent fatty acids in plasma, oils, and tissues.
Agilent 6495C Triple Quadrupole LC/MS
Applied for targeted detection of low-abundance, oxygenated metabolites such as HETEs, DiHETEs, EETs, and prostaglandins. This system offers exceptional sensitivity (LOD < 0.1 ng/mL) and is ideal for studying lipid signaling dynamics.
Agilent 1260 Infinity II HPLC
Employed for chromatographic separation of complex lipid matrices, serving as a front-end module to enhance resolution and reduce matrix interference prior to MS detection.

Agilent 6495C Triple Quadrupole (Figure from Agilent)

Agilent 7890B-5977A (Figure from Agilent)

Agilent 1260 Infinity II HPLC (Fig from Agilent)
Analytical Performance Specifications
| Parameter | Specification |
|---|---|
| Detection Limit (LOD) | < 0.05 ng/mL for oxylipins (e.g., HETEs, DiHETEs) via LC-MS/MS |
| Quantification Range | 0.1 ng/mL – 10 µg/mL (matrix-dependent) |
| Dynamic Range (linear) | ≥ 5 orders of magnitude |
| Accuracy (recovery rate) | 85–115% with internal standard correction |
| Reproducibility (intra-assay CV) | < 10% for most analytes across technical replicates |
| Internal Standards | Stable isotope-labeled analogs (e.g., D₄-AA, D₄-LA) |
| Analyte Coverage | > 40 omega-6 fatty acids and metabolites, including oxylipins |
Sample Requirements for Omega-6 Fatty Acids Analysis Service
| Sample Type | Minimum Amount | Recommended Amount | Storage Condition | Notes |
|---|---|---|---|---|
| Plasma / Serum | 100 µL | ≥200 µL | -80°C, ship on dry ice | Avoid repeated freeze-thaw cycles |
| Tissue (e.g., liver, brain, fat) | 20 mg | ≥50 mg | -80°C, ship on dry ice | Snap-freeze in liquid nitrogen immediately after dissection |
| Cultured Cells | 1×10⁶ cells | ≥5×10⁶ cells | -80°C, ship on dry ice | Cell pellets washed with PBS and flash-frozen |
| Plant Oil / Extract | 100 µL | ≥500 µL | 4°C or ambient (sealed) | Store in amber vials to avoid oxidation |
| Food / Feed / Powder | 0.2 g | ≥1 g | -20°C or ambient (dry) | Please provide detailed matrix information |
| Cell Culture Medium / Supernatant | 200 µL | ≥500 µL | -80°C, ship on dry ice | Filtered or centrifuged to remove debris |
Demo Results
FAQ of Omega-6 Fatty Acids Analysis Service
Can I submit different types of samples within the same project batch?
Yes. We can accommodate mixed sample types (e.g., serum + tissue + oil) in one batch. Each sample will be processed with matrix-specific extraction and analysis methods. Please indicate sample type clearly in your submission form.
Can I customize the list of target compounds to focus on specific fatty acids or metabolites?
Yes. Our service supports both standard and customizable target panels. If you have specific analytes of interest (e.g., 15-HETE, EpOME, or DiHOME), we can adjust the method parameters to include or emphasize these compounds.
Can I compare my omega-6 data with previous batches or external datasets?
We ensure inter-batch consistency through internal standards and QC normalization. If you plan to integrate current data with historical results, please inform us in advance so we can apply consistent methods and calibration.
Do you provide assistance with biological interpretation or pathway mapping?
Yes. In addition to raw and processed data, we offer optional bioinformatics support, including mapping omega-6 metabolites to KEGG or HMDB pathways, lipid class distribution summaries, and basic statistical interpretations (e.g., fold change, z-score).
What quality control measures are included in the analysis?
Each batch includes method blanks, pooled QC samples, and internal standard monitoring. We also track retention time shifts, signal response linearity, and matrix effects to ensure analytical reliability and traceability.
How do you handle low-concentration or borderline-detectable fatty acids?
We apply signal-to-noise ratio thresholds (typically S/N > 3 for detection, >10 for quantitation) and flag low-abundance results accordingly. Compounds below reliable detection limits are clearly marked in the report.
What if my samples contain antioxidants, preservatives, or other additives?
Please notify us in advance. Certain additives (e.g., BHT, EDTA) can interfere with derivatization or ionization. Our technical team can assess compatibility and adjust the protocol or recommend alternative handling.
Do you archive extracted samples or raw materials in case reanalysis is needed?
Yes. By default, we store extracted samples and raw materials at –80°C for 1 month post-analysis. If longer retention is required, please request it in your project form.
Learn about other Q&A.
Omega-6 Fatty Acids Analysis Service Case Study

Title: Simultaneous Quantitative Profiling of 20 Isoprostanoids from Omega-3 and Omega-6 Polyunsaturated Fatty Acids by LC-MS/MS in Various Biological Samples
Journal: Analytica Chimica Acta
Published: 2016
- Background
- Methods
- Results
- Reference
Isoprostanoids are non-enzymatic oxygenated derivatives of polyunsaturated fatty acids (PUFAs), classified as oxylipins. These lipid mediators are increasingly recognized for their roles in oxidative stress, inflammation, vascular integrity, and immune modulation. Among them, F2-isoprostanes (from arachidonic acid), F3-isoprostanes (from EPA), F4-neuroprostanes (from DHA), and F1-phytoprostanes (from ALA) have garnered interest as potential biomarkers of lipid peroxidation and dietary PUFA metabolism.
Due to their structural similarity, low abundance (picogram–nanogram levels), and diversity across PUFA origins, simultaneous profiling of multiple isoprostanoid species in biological samples poses significant analytical challenges. An accurate, sensitive, and reproducible method is critical for elucidating their biological relevance and their modulation by dietary or pathological conditions.
The study developed and validated a robust LC-MS/MS method for the simultaneous quantification of 20 isoprostanoids in various biological matrices, including plasma, cerebrospinal fluid (CSF), urine, liver, brain, and muscle tissue. Key steps included:
- Lipid extraction using modified Folch method with BHT antioxidant
- Base hydrolysis (KOH in methanol) to release esterified isoprostanoids
- Solid-phase extraction (SPE) using Oasis MAX cartridges for sample cleanup
- LC-MS/MS quantification using Agilent 1290 Infinity LC and 6460 Triple Quad MS
- Non-derivatized analysis in negative ion mode
- Method optimized for 20 isoprostanoid standards with validated SRM transitions
- LODs ranged from 0.24–15.6 ng/mL, and LOQs from 0.49–31.25 ng/mL
Although derivatization (e.g., with 2-picolylamine) was explored to improve ionization, it introduced matrix noise and peak overlap, making non-derivatized analysis more suitable for biological samples.
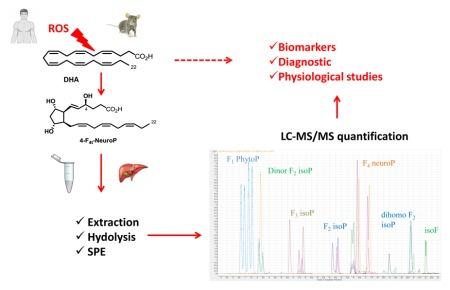 Quantitative LC-MS/MS profiling of 20 isoprostanoids from omega-3 and omega-6 PUFAs.
Quantitative LC-MS/MS profiling of 20 isoprostanoids from omega-3 and omega-6 PUFAs.
Powered by Creative Proteomics
We provide LC-MS/MS-based lipid peroxidation profiling for simultaneous quantification of omega-3 and omega-6-derived isoprostanoids.
- Validated extraction & SPE protocols
- High-sensitivity SRM quantification (LOD < 1 ng/mL)
- Supports plasma, tissue, and nutritional samples
- Custom method development and data interpretation available
The method demonstrated high linearity, precision (RSD ≤ 8%), and inter-day reproducibility (accuracy 81–115%).
Validation in plasma samples showed acceptable matrix effect (51–93%) and extraction yield (40–85%).
The platform successfully profiled isoprostanoids in multiple sample types, with clear tissue-specific distribution of F2-isoprostane and F4-neuroprostane isomers.
DHA supplementation in LDLR−/− mice resulted in:
- A significant increase in total F4-neuroprostanes in liver and brain
- A marked reduction in F2-isoprostanes, particularly in liver tissue
- Strong correlations between DHA levels and F4-NeuroP formation in the liver (R² = 0.96)
The data underscored the plasticity of liver tissue in response to PUFA modulation and emphasized the necessity of comprehensive isoprostanoid profiling for interpreting oxidative stress and lipid remodeling across tissues.
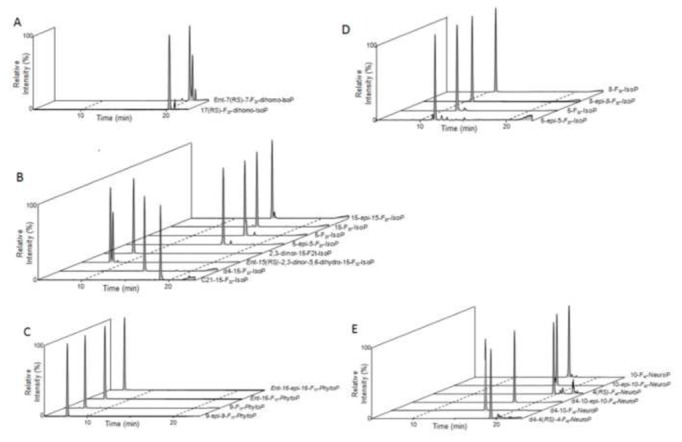 Chromatogram of selected reaction monitoring (SRM) of metabolites from each PUFA A: adrenic acid, B: arachidonic acid, C: α-linolenic acid, D: eicosapentaenoic acid and E: docosahexaenoic acid.
Chromatogram of selected reaction monitoring (SRM) of metabolites from each PUFA A: adrenic acid, B: arachidonic acid, C: α-linolenic acid, D: eicosapentaenoic acid and E: docosahexaenoic acid.
Reference
- Dupuy, Aude, et al. "Simultaneous quantitative profiling of 20 isoprostanoids from omega-3 and omega-6 polyunsaturated fatty acids by LC–MS/MS in various biological samples." Analytica Chimica Acta 921 (2016): 46-58. https://doi.org/10.1016/j.aca.2016.03.024
Publications
Here are some of lipidomics-related papers published by our clients:

- White matter lipid alterations during aging in the rhesus monkey brain. 2024. https://doi.org/10.1007/s11357-024-01353-3
- Characterization of Dnajc12 knockout mice, a model of hypodopaminergia. 2024. https://doi.org/10.1101/2024.07.06.602343
- Evidence for phosphate-dependent control of symbiont cell division in the model anemone Exaiptasia diaphana. 2024. https://doi.org/10.1128/mbio.01059-24
- The olfactory receptor Olfr78 promotes differentiation of enterochromaffin cells in the mouse colon. 2024. https://doi.org/10.1038/s44319-023-00013-5
- Annexin A2 modulates phospholipid membrane composition upstream of Arp2 to control angiogenic sprout initiation. 2023. https://doi.org/10.1096/fj.202201088R
- Lipid Membrane Engineering for Biotechnology. 2023. https://doi.org/10.48780/publications.aston.ac.uk.00046663














