Branched Chain Amino Acids Analysis Service
Struggling with inconsistent BCAA results or incomplete pathway readouts?
Leucine/isoleucine co-elution, matrix interference, and missing intermediates often limit insight into mitochondrial health, nutrient signaling, and cellular metabolism.
At Creative Proteomics, our BCAA analysis service resolves these issues with isomer-specific LC–MS/MS methods and pathway-informed panels. We quantify leucine, isoleucine, valine, their keto acids (BCKAs), hydroxyacids, and acylcarnitines—revealing flux through transamination, β-oxidation, and detox conjugation.
Why Researchers Trust Our BCAA Profiling
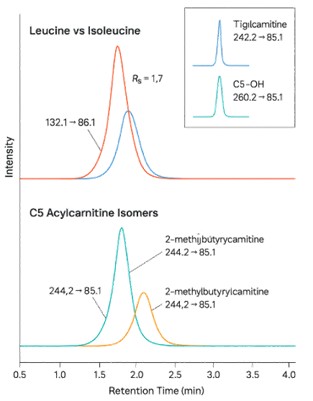
- Isomer-resolved results — Clear separation of leucine vs isoleucine and C5 acylcarnitine isomers
- Matrix versatility — Consistent data from plasma, serum, urine, cells, tissues, and media
- Pathway context — Integrated quantification of BCAAs, BCKAs, hydroxyacids, and conjugates
- High sensitivity, low carryover — Trace detection with verified QC metrics
Submit Your Request Now
×
- What We Provide
- Advantages
- Technology Platform
- Sample Requirements
- Demo
- FAQs
What Are Branched Chain Amino Acids (BCAAs)?
Branched chain amino acids—leucine, isoleucine, and valine—are essential amino acids with branched side chains that influence protein synthesis, cellular energy balance, and metabolic signaling. In research workflows, branched chain amino acids analysis supports pathway mapping and mechanism studies across biofluids, cells, and tissues.
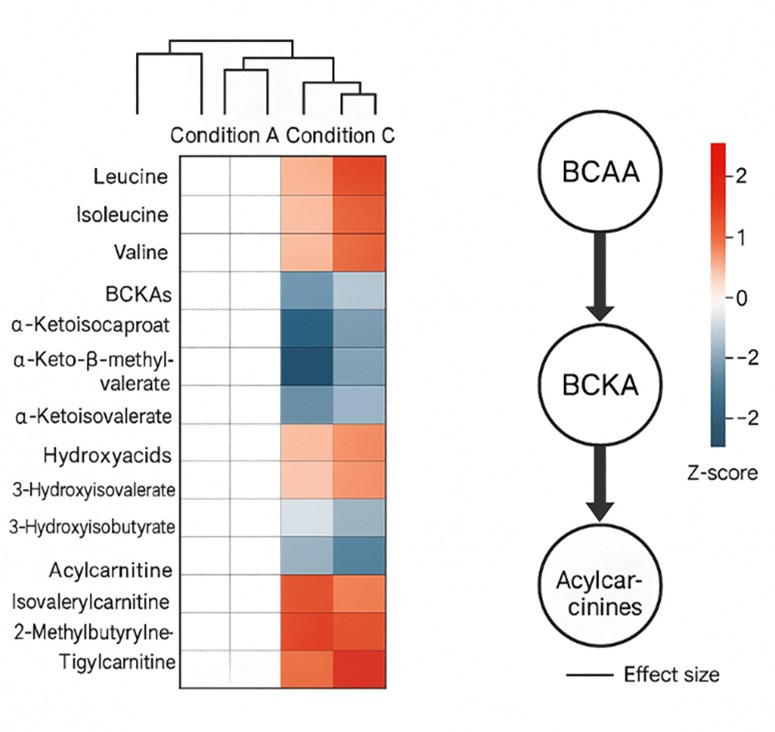
BCAA catabolism begins with transamination to branched-chain keto acids, followed by mitochondrial oxidation that links to the TCA cycle. Tracking parent amino acids alongside acylcarnitines and ketoacid intermediates enables BCAA profiling that clarifies flux, mitochondrial function, and nutrient response in model systems.
What Problems Does This Service Solve?
Most BCAA studies fail on separation, stability, and context. Our service fixes that.
- Isomer resolution: We separate leucine from isoleucine and resolve isobaric C5 acylcarnitines.
- Matrix effects: Method design minimizes ion suppression in plasma, media, urine, and tissues.
- Labile intermediates: Stabilized prep preserves ketoacids and hydroxyacids for reliable readouts.
- Limited dynamic range: Optimized calibration expands coverage from trace to high-abundance levels.
- Batch drift and carryover: Scheduled acquisition and guarded workflows reduce drift and ghost peaks.
- Derivatization variability: Controlled chemistries improve repeatability for amino acid quantification.
- Fragmentation ambiguity: Orthogonal confirmation (e.g., PRM) clarifies identity when MRM overlaps.
- Missing pathway context: Integrated panels link BCAAs with acylcarnitines and ketoacids for flux insight.
BCAA Metabolite Profiling: Service Overview
- Core BCAA Quantification (Leu/Ile/Val): Absolute or relative levels across biofluids, cells, and tissues for baseline and intervention studies.
- Branched-Chain Keto Acids (BCKAs): KIC, KMV, and KIV to map transamination and identify upstream bottlenecks.
- Hydroxyacid Intermediates: 3-hydroxyisovalerate, 3-hydroxyisobutyrate, and related species to assess downstream oxidation.
- Short-Chain Acylcarnitines Panel: C3, C4-iso, C5, C5:1, C5-OH to infer mitochondrial handling and β-oxidation coupling.
- Glycine Conjugates: Isovalerylglycine, 2-methylbutyrylglycine, isobutyrylglycine as detoxification readouts.
- Expanded Context Amino Acids (Optional): Alanine, glutamine, glutamate, and urea-cycle markers for pathway context.
- Stable Isotope BCAA Flux: 13C/15N labeling to quantify uptake, dilution, and oxidative fate with natural-abundance correction.
- Cell Culture Media Monitoring: In-media BCAA consumption/accumulation tracking for process optimization.
- Compound/Condition Screening: Comparative BCAA pathway profiling to prioritize mechanisms of action in discovery settings.
Detectable Analytes for Branched-Chain Amino Acid (BCAA) Profiling
| Class | Analyte (Common Name) | Synonym / Notes |
|---|---|---|
| Core BCAAs | Leucine | Essential BCAA; mTOR-linked signaling |
| Isoleucine | Structural isomer of leucine; requires chromatographic resolution | |
| Valine | Essential BCAA; precursor to propionyl-CoA | |
| Branched-Chain Keto Acids (BCKAs) | α-Ketoisocaproate | KIC; leucine transamination product |
| α-Keto-β-methylvalerate | KMV; isoleucine transamination product | |
| α-Ketoisovalerate | KIV; valine transamination product | |
| Hydroxyacid Intermediates | 3-Hydroxyisovalerate | Leucine oxidative intermediate |
| 3-Hydroxyisobutyrate | Valine oxidative intermediate | |
| 2-Hydroxy-3-methylvalerate | Isoleucine-linked hydroxyacid | |
| Short-Chain Acylcarnitines | Isovalerylcarnitine (C5) | Leucine β-oxidation readout |
| 2-Methylbutyrylcarnitine (C5) | Isoleucine β-oxidation readout | |
| Tiglylcarnitine (C5:1) | Enoyl intermediate in BCAA catabolism | |
| 3-Hydroxyisovalerylcarnitine (C5-OH) | Hydroxyacyl intermediate (leucine) | |
| Isobutyrylcarnitine (C4-iso) | Valine-linked acylcarnitine | |
| Propionylcarnitine (C3) | Downstream of valine; TCA anaplerosis context | |
| Acetylcarnitine (C2) | Global acetyl-flux context (optional) | |
| Glycine Conjugates | Isovalerylglycine | Leucine pathway detox marker |
| 2-Methylbutyrylglycine | Isoleucine pathway detox marker | |
| Isobutyrylglycine | Valine pathway detox marker | |
| Context Amino Acids (Optional Panel) | Alanine | Transamination partner; redox context |
| Glutamine / Glutamate | Nitrogen shuttle; anaplerosis indicators | |
| Citrulline / Ornithine | Urea-cycle linkage | |
| Phenylalanine / Tyrosine | Amino acid balance references |
CoA thioesters are generally inferred via validated proxies under RUO conditions. Custom targets may be added following method feasibility review.
Why Choose Creative Proteomics for Branched Chain Amino Acid Analysis?
- Isomer-true results you can act on: Baseline resolution of leucine vs isoleucine and separation of C5 acylcarnitine isomers (isovaleryl vs 2-methylbutyryl) to avoid pathway misreads.
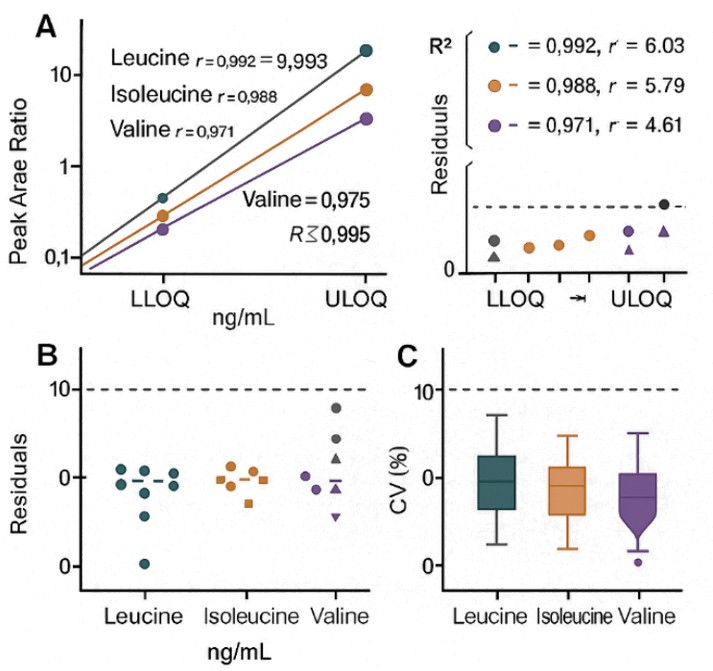
- QC metrics reported transparently: Multi-point calibration with isotope-labeled internal standards; typical fits R2 ≥ 0.995 and method precision CV ≤ 10% for core BCAAs, disclosed in your report.
- Confidence at low levels: Guarded wash and injection logic designed for<0.1% LLOQ carryover on target transitions, supporting trace-level detection without memory effects.
- Tracer-ready analytics (RUO): Built-in natural-abundance and isotope-interference correction for ^13C/^15N studies, plus flux-style tables that link parent BCAAs to BCKAs and acylcarnitines.
- Matrix-agnostic robustness: Protein precipitation/SPE options and optional derivatization reduce ion suppression across plasma/serum, tissues, cells, media, and urine, keeping quantitation stable.
- One report, full context: Parent amino acids, BCKAs, hydroxyacids, and acylcarnitines delivered as an integrated panel with pathway visuals, so mechanism signals are clear at a glance.
- Sample-sparing by design: High-sensitivity methods support low-volume injections, helping preserve limited or precious research materials.
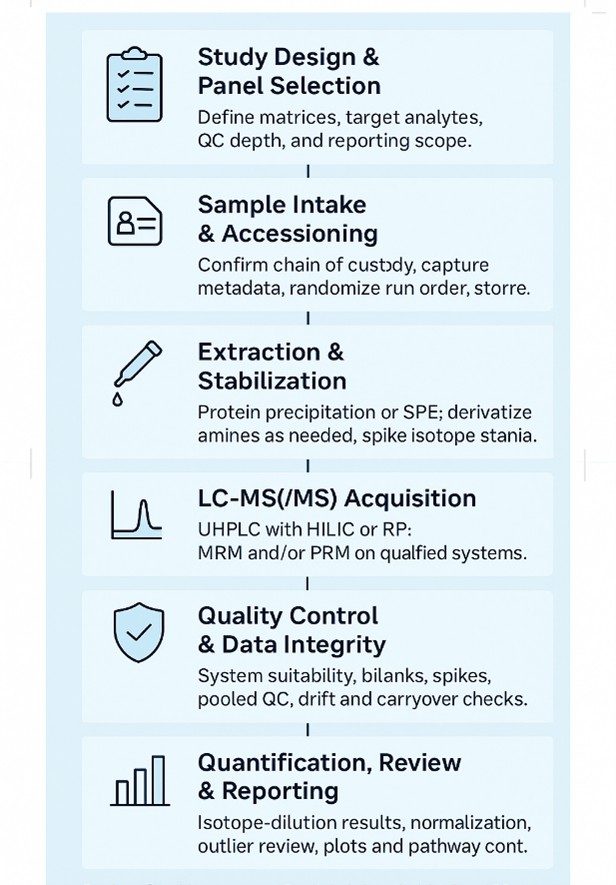
Branched Chain Amino Acid Analysis Workflow: Step-by-Step Process
- Study Design & Panel Selection — Define matrices, target analytes, and reporting needs; choose core or extended panels.
- Sample Receipt & Logging — Verify chain of custody, record metadata, and store under controlled conditions with randomized run order.
- Extraction & Stabilization — Perform protein precipitation or SPE; apply optional derivatization for amino acids and add stable-isotope standards.
- LC–MS(/MS) Acquisition — Run UHPLC with HILIC or reversed-phase separation; acquire on triple-quadrupole MRM and/or HRAM PRM.
- Quality Control — Include system suitability, procedural blanks, spikes, pooled QC, matrix-matched calibration, and drift checks.
- Data Processing — Use isotope-dilution quantification, normalization, and outlier review with predefined acceptance rules.
- Reporting & Review — Deliver data tables, plots, and pathway context, with concise interpretations mapped to study objectives.
BCAA Analysis: Core Instruments & Key Parameters
UHPLC Systems (Primary)
- Models: Agilent 1290 Infinity II, Thermo Vanquish
- Separation: HILIC (amide) for native amino acids; Reversed-phase C18 for acylcarnitines/hydroxyacids
- Key params: autosampler cooling; sub-2–3 μm columns; ion-pair–free methods or PITC/OPA/FMOC derivatization when required
Targeted Quantification (Triple Quadrupole)
- Models: AB Sciex QTRAP/Triple Quad 6500+, Agilent 6495C
- Modes: Scheduled MRM, fast polarity switching (ESI +/–)
- Key params: isotope-labeled internal standards; optimized dwell/RT windows for Leu/Ile and C5 isomers
Identity Confirmation / Extended Panels (HRAM)
- Models: Orbitrap Exploris 480, Q Exactive HF-X
- Modes: PRM for isomer verification; Full MS/MS for interference checks
- Key params: resolving power ~60k–120k (m/z 200); lock-mass/RT calibrants for retention stability

1260 Infinity II HPLC (Figure from Agilent)

6495C Triple Quadrupole (Figure from Agilent)

Orbitrap Exploris 480 (Figure from Thermo)
Sample Requirements for BCAA Analysis Service
| Matrix | Reference Amount per Sample | Recommended Container / Additive | Preparation Notes |
|---|---|---|---|
| Plasma / Serum | ≥150–200 µL | Low-bind screw-cap tube; K2-EDTA or heparin for plasma | Avoid hemolysis; record anticoagulant; keep cold before freezing |
| Urine | ≥1 mL | Polypropylene tube; no preservative | Mix gently; aliquot to reduce freeze–thaw cycles |
| Cell Lysate | ≥200–300 µg total protein or ≥200 µL lysate | Low-bind tube; compatible extraction buffer | Note buffer components (salts/detergents); keep on ice pre-freeze |
| Cell Pellet | ≥1–5 × 106 cells | Cryovial; no medium | Wash to remove media; snap-freeze promptly |
| Tissue (Animal/Plant) | ≥30–50 mg wet weight | Foil-wrapped cryovial or tube | Rinse to remove blood/medium when appropriate; snap-freeze |
| Culture / Conditioned Media | ≥500–1,000 µL | Sterile tube or bottle | Record base medium and supplements; clarify culture timepoint/condition |
| CSF (Optional) | ≥200–300 µL | Low-bind tube | Collect with minimal blood contamination |
General guidance:
Use leak-proof, low-bind plastics with clear labels (ID, matrix, additive). Avoid ion-pair reagents or strong acids/bases. Provide a printed manifest listing sample IDs and target analytes.
BCAA Profiling — Demo Results
Case Study: Client Success Stories

Comparative metabolite profiling of salt sensitive Oryza sativa and the halophytic wild rice Oryza coarctata under salt stress
Journal: Plant-Environment Interactions
Published: 2024
- Background
- Methods
- Results
- Conclusions
- Reference
Salinity severely disrupts plant metabolism. The study compares root metabolomes of salt-sensitive Oryza sativa and halophytic Oryza coarctata to pinpoint metabolite classes and pathways linked to salt tolerance. Under non-stress conditions, O. coarctata shows higher abundance for many metabolites; salinity then triggers distinct, species-specific shifts, suggesting different coping strategies.
The authors performed untargeted, root-specific comparative metabolomics across control and salt-stressed plants, followed by multivariate statistics (e.g., PCA, hierarchical clustering), differential analysis (volcano plots, Venn comparisons), lipid-class heatmaps, and KEGG pathway enrichment. Results are reported at chemical super-class and pathway levels to connect metabolite changes with tolerance traits. (Analytical specifics are in the journal; the PubMed record confirms the untargeted comparative design and downstream analyses.)
- Global differentiation: PCA and clustering separate control vs salt in both species, confirming broad metabolic reprogramming.
- Constitutive abundance in O. coarctata: Under control conditions, itaconate, vanillic acid, threonic acid, eicosanoids, xanthine-related compounds, and eight amino acids are comparatively higher, suggesting a pre-tuned metabolic state.
- Lipid remodeling: Glycerolipids and many phospholipids are lower in O. coarctata at baseline and decrease further with salt; fatty acyls increase in O. coarctata, while organic acids are prominently induced in O. sativa.
- Pathway enrichment linked to tolerance: Arachidonic acid metabolism appears upregulated in O. coarctata; phenylpropanoid, cutin/suberin/wax biosynthesis are more enriched, consistent with anatomical traits that impede ion entry and water loss.
 PCA + Heatmap
PCA + Heatmap
PCA separates control and salt-stressed samples for both species; the heatmap (top metabolites) shows coherent clustering by treatment.
 Volcano Plots (Four Comparisons)
Volcano Plots (Four Comparisons)
Differential metabolite profiles per comparison; points colored by direction and significance thresholds.
O. coarctata deploys a distinct metabolic program under salt stress, characterized by constitutive abundance of stress-relevant metabolites, selective induction of fatty acyls, and enrichment of barrier-forming and signaling pathways. These features likely underpin halophytic tolerance and point to candidate metabolic targets for engineering resilience in cultivated rice.
Tamanna, Nishat, et al. "Comparative metabolite profiling of salt sensitive Oryza sativa and the halophytic wild rice Oryza coarctata under salt stress." Plant‐Environment Interactions 5.3 (2024): e10155.
FAQ of BCAA Analysis Service
How do you achieve isomer-true quantitation for leucine vs isoleucine?
Isomer separation is obtained chromatographically rather than inferred from MS alone; hydrophilic interaction chromatography (HILIC) or carefully selected reversed-phase methods provide distinct retention, and orthogonal MS confirmation (e.g., PRM or specialized fragmentation) safeguards identity when transitions overlap.
Can C5 acylcarnitine isomers be resolved (isovaleryl vs 2-methylbutyryl, and related species)?
Yes; validated UHPLC–MS/MS methods resolve C5 isomers to reduce false positives, a practice also adopted in newborn-screening second-tier testing and modern vendor application notes.
What role do branched-chain keto acids (BCKAs) play in interpretation?
BCKAs (KIC, KIV, KMV) are the first transamination products and provide readouts of upstream flux and bottlenecks; multiple LC–MS/MS methods demonstrate their simultaneous measurement alongside BCAAs for pathway context.
Do I need derivatization for amino acid analysis?
Not always; underivatized amino acids can be measured with HILIC–MS, while pre-column derivatization chemistries (e.g., PITC or AccQ-Tag) can enhance sensitivity or chromatographic performance—selection is driven by matrix and targets.
How are matrix effects controlled across plasma, tissues, cells, urine, or media?
Matrix effects are mitigated by matrix-matched calibration, stable-isotope internal standards, and optimized sample prep/derivatization; current best-practice reviews emphasize tailoring derivatization and chromatographic conditions to reduce suppression and improve accuracy.
Can isotope-tracer studies (13C/15N) be integrated into BCAA panels?
Yes; tracer-enabled LC–MS/MS panels quantify labeled enrichment of BCKAs and related metabolites together with endogenous levels, enabling flux-style interpretation when natural-abundance and interference corrections are applied.
What outputs should I expect to interpret results quickly?
Deliverables that speed decisions include annotated chromatograms with retention-time windows, calibration and QC summaries, and data tables linking parent BCAAs to BCKAs and acylcarnitines; such reporting aligns with method papers where simultaneous quantification and QC acceptance criteria are standard.
How do you verify that separation, not fragmentation, drives specificity?
Specificity is evidenced by baseline chromatographic resolution (e.g., leucine vs isoleucine; C5 isomer pairs) plus confirmatory MS behavior; literature and vendor notes document that resolving power and tuned methods lower false identifications compared with MS-only strategies.
Learn about other Q&A about proteomics technology.
Publications
Here are some publications in Metabolomics research from our clients:

- Non-invasive elevation of circulating corticosterone increases the rejection of foreign eggs in female American robins (Turdus migratorius). 2022. https://doi.org/10.1016/j.yhbeh.2022.105278
- Polyamine metabolism impacts T cell dysfunction in the oral mucosa of people living with HIV. 2023. https://doi.org/10.1038/s41467-023-36163-2
- Overexpression of maize ZmLOX6 in Arabidopsis thaliana enhances damage-induced pentyl leaf volatile emissions that affect plant growth and interaction with aphids. 2023. https://doi.org/10.1093/jxb/erac522
- Proteolytic activation of fatty acid synthase signals pan-stress resolution. 2024. https://doi.org/10.1038/s42255-023-00939-z
- Nutritional analysis of commercially available, complete plant-and meat-based dry dog foods in the UK. 2024. https://doi.org/10.1101/2024.09.11.612409