Proteomics has become an increasingly important tool in Alzheimer's disease research, providing valuable insights into the underlying mechanisms and progression of the disease. By studying the changes in protein expression and modifications, researchers are able to gain a better understanding of the molecular changes that occur in the brain as Alzheimer's progresses. Proteomics can also be used to identify new biomarkers and drug targets, which can be used to develop new treatments and diagnostic tools. For example, by analyzing the cerebrospinal fluid of patients with Alzheimer's, researchers can identify specific protein changes that are indicative of the disease, which can be used as early markers for diagnosis. Additionally, proteomics can be used to evaluate the efficacy of potential therapeutic strategies by measuring changes in protein expression and modifications in response to treatment. Overall, proteomics plays a crucial role in advancing our understanding of Alzheimer's disease and developing new strategies for prevention, diagnosis, and treatment.
Case 1 Proteomic screening for signature proteins in early AD (1)
The ε4 allele of the apolipoprotein E (APOE) gene is the most risky genetic factor for sporadic or late-onset Alzheimer's disease (AD), but the exact mechanism mediating the risk of APOE ε4-associated Alzheimer's disease is unknown, and APOE-based treatments for Alzheimer's disease have not yet been achieved.
The authors compared the protein expression of the temporal lobe (ITG) and middle frontal gyrus (MFG) in individuals with Alzheimer's disease (AD) and control groups in the ROS and BLSA cohorts. In the BLSA cohort, 0 and 254 different proteins were identified in the ITG and MFG, respectively. In the ROS cohort, 244 and 4 different proteins were identified in the ITG and MFG, respectively. The intersection of the BLSA and ROS cohorts resulted in 120 proteins, of which 84 were downregulated and 34 were upregulated.
The intersection of the 120 proteins that showed differences in both ROS and BLSA cohorts and the YAPS cohort (early AD) with APOE ε4+ versus APOE ε4- resulted in 16 and 14 common different proteins in the ITG and MFG, respectively, a total of 25 unique proteins. These proteins were further analyzed and confirmed to be characteristic proteins of early AD patients. Pathway enrichment analysis showed that these 25 characteristic proteins were enriched in cytokine signaling, tyrosine kinase signaling, cell migration, and other signaling pathways. Protein-protein interaction analysis (PPI) found that STAT3, FYN, and YES1 were central node proteins.
Further analysis revealed that the proteomic features of early AD patients are related to severe AD pathology and related to cognitive performance prior to developing AD. The authors depicted the proteomic features of AD by analyzing and comparing the proteomic results of brain tissue from Alzheimer's disease patients in different cohorts and identified potential targets for early AD treatment in further experiments.
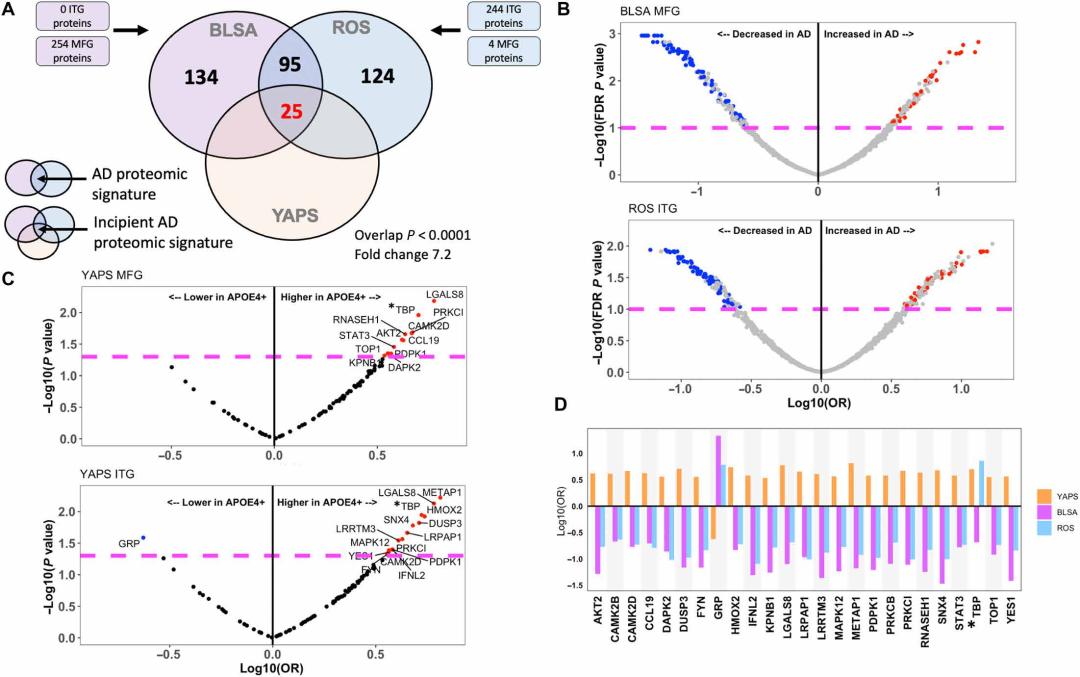
Identifying an incipient AD proteomic signature.
Case 2 Large-scale deep multi-layer analysis of Alzheimer's disease brain (2)
In this study, the authors analysed a total of 516 dorsolateral prefrontal cortex (DLPFC) tissues from control, asymptomatic AD (AsymAD) and AD brains using TMT-MS quantitative proteomics. After data processing and removal of outliers, a total of 8619 proteins were included using the Weighted Gene Co-expression Network Analysis algorithm (WGCNA) to construct a protein co-expression network containing 44 modules and delineate their biological functions using GO analysis.
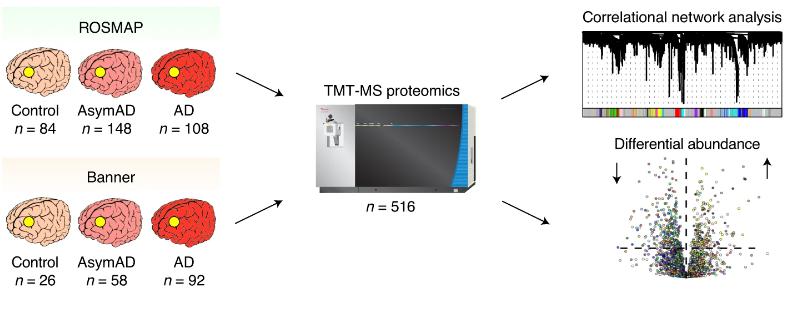
TMT AD protein co-expression network.
The authors observed that a large proportion of these 44 modules were significantly associated with at least one of the pathological manifestations of AD or with cognitive function. Twelve modules, including postsynaptic density, glycosylation/endoplasmic reticulum (ER), oligodendrocyte/myelination, RNA splicing, matrisome, cell-ECM interactions, synapse/neuron, ubiquitination, mitogen-activated protein kinase (MAPK) signaling and metabolism, mitochondria, sugar metabolism, and protein transport, were more strongly associated with AD than others.
The authors focused specifically on two AD-related modules in the proteome: one associated with MAPK signalling pathway and metabolism and one associated with the matrisome, which refers to a collection of extracellular matrix (ECM)-associated proteins. The matrisome module was also found to be influenced by the APOE ε4 allele, but after adjusting for neuropathology it was found to be unrelated to the rate of cognitive decline, whereas the MAPK/metabolic module was strongly associated with the rate of cognitive decline.
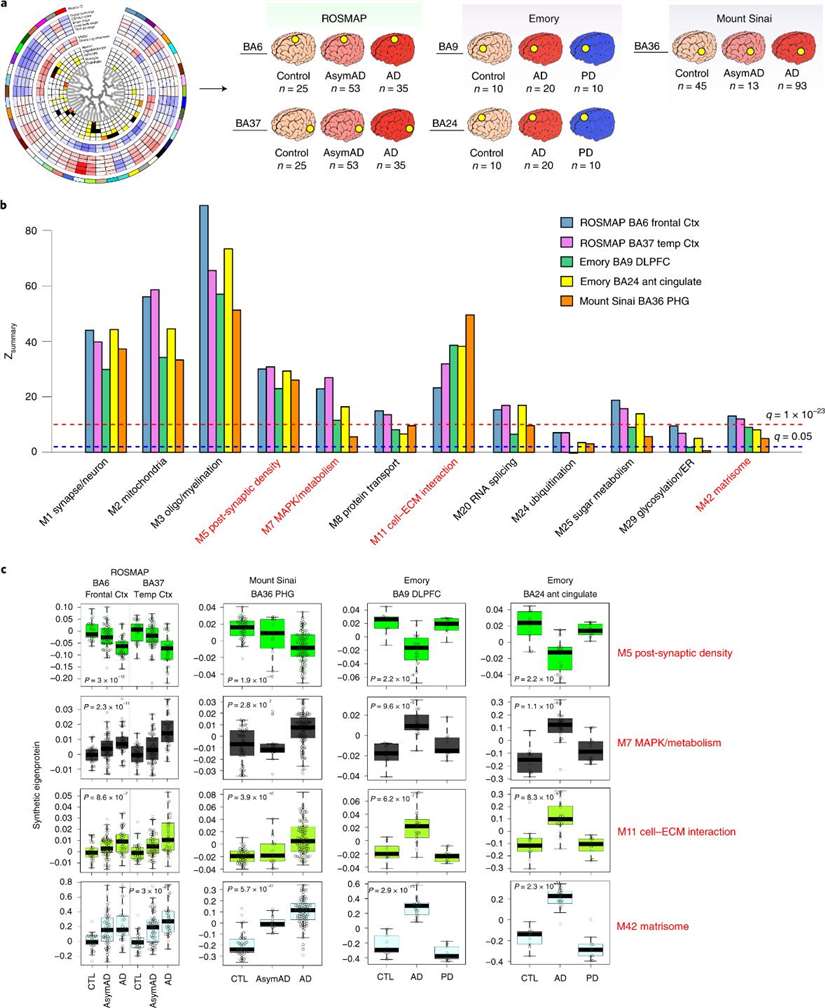
Preservation of the TMT AD network across different cohorts, centers, methods and brain regions.
Dissecting disease-associated proteomics modules may provide reliable biomarkers for AD and offer hope for treatment. And combined multi-omics analysis will undoubtedly provide a better understanding of the relationships between these different data types in the context of AD.
Reference
- Roberts, Jackson A., et al. "A brain proteomic signature of incipient Alzheimer's disease in young APOE ε4 carriers identifies novel drug targets." Science advances 7.46 (2021): eabi8178.
- Johnson, Erik CB, et al. "Large-scale deep multi-layer analysis of Alzheimer's disease brain reveals strong proteomic disease-related changes not observed at the RNA level." Nature neuroscience 25.2 (2022): 213-225.






