1 Proteome and Peptide Preparation
a) Disrupt 25 million pelleted HegG2 cells in the dark in 5 mL of cell disruption buffer for 5 min at room temperature and transfer on ice for 30 min. To ensure adequate cell lysis, repeatedly vortex this solution.
b) Centrifuge for 10 min at 13,000 × g at 4°C and discard the pellet containing insoluble material.
c) Desalt the protein solution on two PD-10 columns (2.5-mL protein solution per column) and collect each desalted protein fraction in 3.5 mL of protein digestion buffer.
d) Mix the two desalted protein solutions together.
e) Add 40 μg of trypsin and incubate overnight at 37°C.
f) Adjust the pH of the peptide solution to pH 3 by adding 1 M HCl.
g) Remove insolubilities by centrifugation for 10 min at 13,000 × g.
h) Load phosphopeptides on Fe3+-loaded IMAC beads according to the manufacturer's protocol.
i) Elute phosphorylated peptides from the IMAC resin with 2 mL of 0.4 M ammonium hydroxide.
j) Split the peptide mixture in two equal parts (mixtures A and B) and dry to complete dryness in a centrifugal vacuum concentrator.
k) Re-dissolve mixture A in 50 μL of calf intestinal alkaline phosphatase (CIP) reaction buffer and add 10 units of CIP, 200 units of lambda protein phosphatase and 0.66 units of alkaline Escherichia coli phosphatase. Incubate for 1 h at 37°C and lower the pH to 5 by adding 80 μL 0.5 M KH2PO4. Add 4 μg sequencing-grade modified trypsin and dry to complete dryness. Redissolve mixture A in 150 μL H218O and incubate overnight at 37°C. Transfer this 18O-labeled, dephosphorylated peptide mixture to an Eppendorf tube containing 1.5 μmol tris(2-carboxyethyl) phosphine (TCEP) and 15 μmol iodoacetamide both present as a dried pellet. Incubate for 1 h at 37°C in the dark and store at −20°C until further use.
l) Repeat step 11 for mixture B but exclude phosphatases and replace H218O by natural water.
2 COFRADIC Isolation of Ex-Phosphorylated Peptides
a) Combine peptide mixtures A and B.
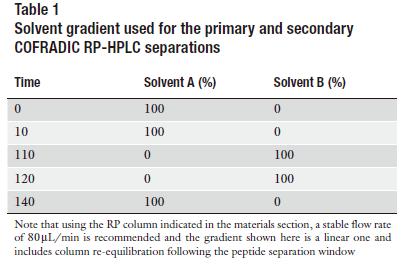
b) Fractionate the peptides by RP-HPLC using the gradient depicted in Table 1 and collect primary fractions using the scheme shown in Table 2.
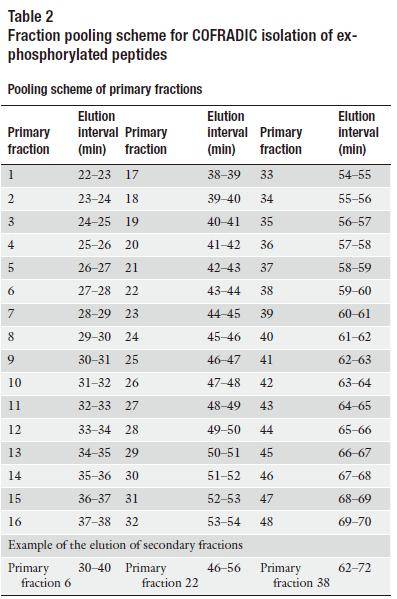
c) Combine primary fractions separated by 16 min (see Table 2), dry and re-dissolve them in 50 μl of CIP reaction buffer containing 10 units of CIP, 200 units of lambda protein phosphatase and 0.66 units of alkaline Escherichia coli phosphatase.
d) Dephosphorylate for 1 h at 37°C and stop the reaction by adding 40 μL of HPLC solvent A and 10 μL of 50 mM acetic acid.
e) Refractionate each pool of dephosphorylated peptides individually on the same column and same solvent gradient (Table 1) as used for the primary RP-HPLC run and collect sorted, ex-phosphorylated peptides (hydrophobic shift) using the sorting scheme shown in Table 2.
f) 6. Store sorted peptides at −20°C until further analysis.
Reference
- de Graauw, M. (2009). Phospho-Proteomics. Humana Press.





