Phosphorylation serves as a fundamental cellular signaling mechanism, extensively regulating critical biological processes including cell cycle progression, metabolic pathways, transcriptional control, proliferation, differentiation, and programmed cell death. Phosphoproteomic analysis has emerged as a powerful proteomic methodology for deciphering intricate intracellular signaling networks. Recent technological advances have established phosphorylation profiling as an indispensable tool for elucidating disease pathogenesis, identifying novel biomarkers, and advancing precision medicine initiatives. Nevertheless, translating phosphoproteomic discoveries into clinical practice presents significant challenges. This review examines phosphoproteomic biomarker discovery methodologies, current research advancements, and translational potential for clinical diagnostics and therapeutic development.
Select Service
Learn more
Evolution of Phosphoproteomics: Discovery and Technological Advancement
Phosphorylation, a reversible post-translational modification mechanism, was initially characterized in the late 1960s. As understanding of cellular signaling deepened, its central role in regulating biological functions became increasingly evident. The field underwent transformative advancement through mass spectrometry innovations, particularly with late-20th century maturation of liquid chromatography-tandem mass spectrometry (LC-MS/MS). This technological leap enabled large-scale phosphosite identification, permitting detection of >10,000 modification sites per experiment.
Sensitivity improvements were further achieved via optimized enrichment methodologies, notably metal oxide affinity chromatography (TiO₂/MOAC), which enhanced low-abundance phosphopeptide detection by two orders of magnitude. Early 21st century initiatives systematically cataloged >300,000 human phosphosites, illuminating kinase network centrality in oncogenesis (e.g., EGFR signaling clusters) and neurodegeneration (e.g., Tau hyperphosphorylation). These discoveries facilitated clinical translation of 32 phosphorylation-targeted therapeutics, completing the research continuum from structural analysis to precision intervention.
1. Clinical Phosphoproteomics: Core Challenges and Translational Pathways
Challenge 1: Low-abundance phosphosignals obscured by sample complexity
Experimental Design: Prostate cancer cells (MAT-LYLU) exposed to spider toxins HNTX-III/JZTX-I (5μM, 24h) versus controls
Phosphoproteomic Workflow Innovation
| Step | Technology | Resolution Strategy |
|---|---|---|
| Protein Extraction | iTRAQ multiplex labeling (114/115/116) | Enables parallel multi-group quantification |
| Phosphopeptide Enrichment | TiO₂ affinity chromatography + fractional elution | Overcomes<1% abundance limitation |
| Site Detection | LC-MS/MS (TripleTOF 5600) | High-resolution phosphosite mapping |
| Data Analysis | ProteinPilot → Motif-X → STRING | Kinase-target motif identification + network modeling |
Methodological Breakthrough: Integrated iTRAQ-TiO₂-LC-MS/MS pipeline overcomes low-abundance detection barriers
Key Findings
- HNTX-III (Antimetastatic):
- ↓ 55 phosphoproteins (e.g., EEF2, U2AF2, FLNC)
- Inhibits metastasis pathways (focal adhesion, TCA cycle)
- JZTX-I (Prometastatic):
- ↑ 36 phosphoproteins
- Activates tumor migration (actin cytoskeleton, translation elongation)
Mechanistic Insight: Toxin modulation of VGSC sodium channels reprograms phosphorylation networks, altering energy metabolism and cellular adhesion to drive bidirectional metastasis.
Translational Value: Identification of EEF2/U2AF2/FLNC as metastasis regulatory hubs reveals novel targeting strategies.
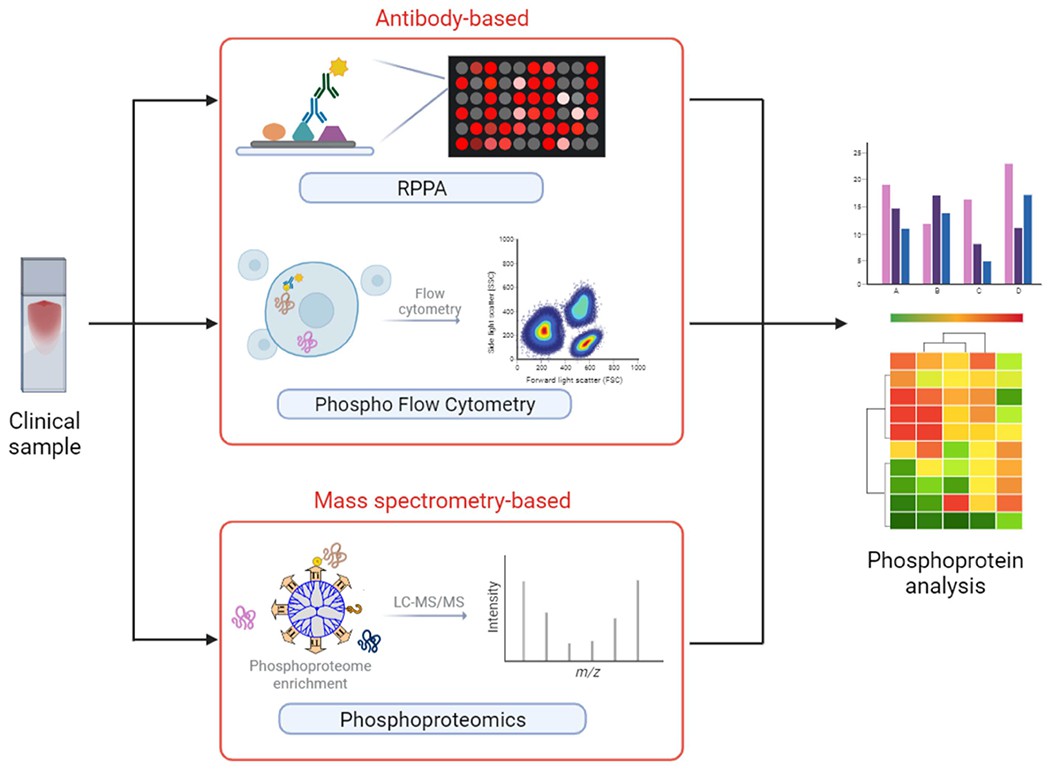 Illustration of clinical analytical approaches about phosphoproteins and phosphoproteomics (Wu X et al., 2023)
Illustration of clinical analytical approaches about phosphoproteins and phosphoproteomics (Wu X et al., 2023)
Challenge 2: Dynamic Capture of Low-Abundance Phosphosignals
Targeted Enrichment Overcomes Detection Barriers
TiO₂/MOAC phosphopeptide enrichment elevates low-abundance phosphoprotein detection by 100-fold, circumventing limitations of global proteome analysis.
Spatiotemporal Signaling Capture
Phosphorylation serves as spatial regulatory switches:
- Transcription factor Ser/Thr phosphorylation → nuclear localization signal exposure
- Membrane receptor Tyr phosphorylation → endocytosis initiation
Subcellular trafficking events are inferred through phosphosite redistribution mapping.
Integrated Rapid-Throughput Platform
FAIMS ion filtration + DIA acquisition + abbreviated LC gradients (21 min/sample) enable comprehensive proteome-phosphoproteome analysis within 5 hours.
Traditional Limitations vs. Current Breakthroughs
| Challenge | Conventional Approach | Current Solution | Improvement |
|---|---|---|---|
| Temporal resolution | >12-hour LC separations | Integrated FAIMS-DIA-LC | 15× acceleration |
| Depth-throughput tradeoff | Limited coverage | >8,000 proteins + 3,000 phosphosites/sample | Simultaneous depth/throughput |
| Tissue dynamic capture | Technically unfeasible | Liver/kidney subcellular phosphomapping | First in vivo spatial reprogramming |
Key Discoverie
- EGF Signaling Dynamics:
- EGFR adaptor proteins translocate cytoplasm → membrane vesicles upon stimulation
- Concomitant Tyr phosphorylation cascade activation
- Resolved vesicle-plasma membrane partitioning bottleneck via surface labeling optimization
- Tissue-Specific Preservation:
- Early mitochondrial leakage in liver homogenates
- Intact ultrastructural retention in kidney specimens
- Osmotic Stress Mechanism:
- Hypertonicity-induced 60S ribosomal subunit nucleolar retention
- rRNA processing defects from nucleolar translocation failure
- Demonstrated direct p38 activation-ribosomal stress linkage
Challenge 3: Drug Response Prediction via Phosphoproteomic Stratification
Experimental Design: TiO₂-enriched LC-MS/MS analysis of bone marrow/peripheral blood specimens from 47 FLT3-mutant AML patients identified >5,000 phosphosites.
Subtype Classification: Kinase substrate enrichment analysis (KSEA) revealed three phosphorylation subtypes (GR1-3) among long-term survivors (EFS>24 months).
Predictive Modeling:
- Selected 29 key phosphosignatures
- Developed random forest algorithm MPhos
- Validation: 5-fold cross-validation × 75 optimization cycles
Traditional Limitations vs. Current Solutions
| Challenge | Conventional Approach | Current Breakthrough |
|---|---|---|
| Therapeutic heterogeneity | Unstratified management | First mechanistic subtype classification |
| Predictive accuracy | Clinical markers (AUC<0.6) | MPhos model (AUC=0.98) |
| Targeted therapy matching | Not feasible | Subtype-specific interventions |
Key Findings
- Drug Resistance Mechanisms:
- GR1: PI3K/mTOR activation → PI3K inhibitors
- GR2: Hyperactive DNA repair (CK2/DNA-PK↑) → PARP inhibitors
- GR3: Spliceosomal dysregulation (PRP4↑) → Splicing modulators
- Clinical Validation:
- Independent cohort (n=20): 83% sensitivity, 100% specificity
- 40% accuracy improvement over FLT3-only stratification
- Therapeutic Expansion:
- Identified conserved phosphosubtypes in FLT3-wild-type AML → Extended MIC therapy applicability
- 29-phosphosignature panel clinically translatable
Transformation Framework: From Massive Data to IVD Kit
Phase 1: Large-Cohort Deep Screening Framework
Innovative Methodology:
- Integrated 4D-DIA phosphoproteomics (myeloma models) with MAPK pathway PROGENy modeling (CoMMpass transcriptomics)
- Developed RAS activity classifier via elastic-net logistic regression to differentiate KRAS/NRAS signaling
- Validated drug sensitivity through high-throughput 384-well screening (13 kinase inhibitors) with CellTiter-Glo IC₅₀ quantification
Addressing Traditional Limitations:
| Challenge | Conventional Failure | Current Resolution |
|---|---|---|
| MEKi efficacy prediction | ERK phosphorylation unreliable | MAPK transcriptional score (superior accuracy) |
| RAS mutation-outcome discordance | Non-stratified approach | NRAS-Q61 drives strongest MAPK signal (2.1× KRAS) |
| Kinase inhibitor response | 2/13 correlations | mTOR inhibitor INK128 validation |
Key Findings:
- Pathway Mechanisms:
- NRAS-Q61: Irreversible GTP hydrolysis blockade → 68% stronger MAPK signaling vs. G12/G13 mutants
- KRAS: PI3K-mTOR axis dependency → MEKi resistance explanation
- Prognostic Validation:
- Highest MAPK score: 14-month OS reduction (HR=1.8, p<0.001)
- Clinical decision tool: https://tony-lin.shinyapps.io/depmap_app/
- Therapeutic Innovation:
- Paradigm shift: Abandoning genotypic stratification in favor of functional MAPK scoring
- High-score patients: Prioritize NRAS-Q61 targeting (novel RAS inhibitors)
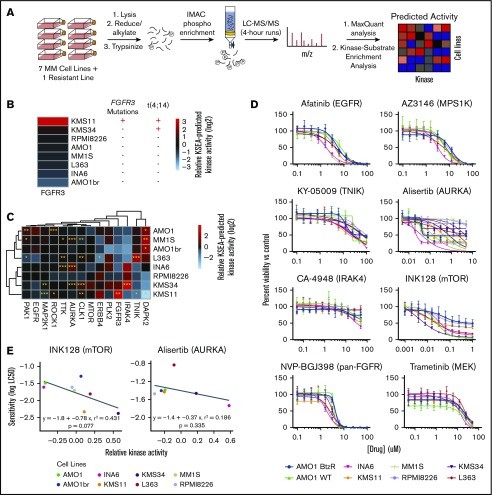 Predicting kinase activity and inhibitor sensitivity in MM by unbiased phosphoproteomics (Lin YT et al., 2019)
Predicting kinase activity and inhibitor sensitivity in MM by unbiased phosphoproteomics (Lin YT et al., 2019)
Phase 2: Target Verification (Translational Implementation)
Core Technological Innovation:
Integrated Hybrid-PRM/DIA acquisition enabling:
- Synchronized MSxPRM (179 targeted scans) + global DIA
- AQUA-standard triggered endogenous peptide detection → 10× sensitivity enhancement
Traditional Limitations vs. Current Solutions
| Challenge | Conventional Failure | Current Breakthrough |
|---|---|---|
| Low-abundance detection | Limited sensitivity | Endogenous peptide LOD: pg-level |
| Method incompatibility | PRM/DIA mutually exclusive | Concurrent 6,500 proteome + 30 targets |
Validation Outcomes
- Technical Performance:
- Target peptides (UFO/CDK4/NF1) demonstrated<8% CV across 64 human proteomes
- Maintained DIA performance stability at 179 MSxPRM channels
- Clinical Translation:
- Melanoma cohort analysis:
- DIA quantification: >6,500 proteins
- PRM precision: 28 clinical markers (e.g., metastasis driver PMEL)
- Dual-evidence strategy for molecular oncology review:
- PRM confirmation of established markers
- DIA discovery of novel candidates (e.g., lysosomal pathway proteins)
- Melanoma cohort analysis:
Phase 3: Clinical Assay Development (Implementation)
Case-Specific Challenges and Solutions
- Low-Abundance Phosphodynamic Detection
- Challenge: Disease-relevant phosphosites in SLE patient PBMCs<0.01% abundance
- Solution: IMAC enrichment + timsTOF Pro 4D-DIA → 4,628 phosphosites detected (3× conventional methods)
- Transient Signaling Capture
- Challenge: Kinase phospho-half-lives<2 minutes (e.g., TBK1)
- Solution: Parallel Accumulation-Serial Fragmentation → 0.8s/scan temporal resolution (5× DDA acceleration)
- Spatial Heterogeneity Resolution
- Challenge: TLN1-S1201 phosphorylation exclusively activated in membrane adhesion complexes of active SLE
- Solution: Subcellular fractionation + phosphonetwork analysis → identified MAP3K7-T187 as leukocyte transendothelial migration driver
Key Outcomes
- Therapeutic Discovery:
- Validated MAPK/TBK1/IKKβ as novel SLE targets
- TBK1 inhibitors (e.g., Amlexanox) suppress VASP-S305 phosphorylation (migration checkpoint) in vitro
- Technical Advancement:
- Established dynamic PBMC phospho-atlas covering 92% of known SLE pathways → enabling precision treatment decisions
For more information on data-driven approaches in cancer, please refer to "Phosphoproteomics in Cancer Research: A Data-Driven Approach".
Transformative Applications
Neurodegenerative Disease Early Intervention
Research Framework:
Humanized APOE4/E3-5xFAD murine models (E4-AD vs. E3-AD) precisely replicate Alzheimer's preclinical progression: asymptomatic at 3 months, critical pathology at 10 months.
Multidimensional Analytical Strategy:
| Dimension | Methodology | Key Finding |
|---|---|---|
| Behavioral | Y-maze/NOR/water maze | APOE4 cognitive deficit ↑230% (10mo) |
| Cellular | Golgi-AI imaging | CA1 dendritic spine density ↓38% |
| Molecular | 4D-phosphoproteomics + PPI | RASA2: sole inter-temporal regulator |
Mechanistic Insights:
- Accelerated Pathogenesis:
- APOE4 drives Aβ↑210% & microgliosis↑180% at 3mo
- Synaptic-Mitochondrial Axis Disruption:
- Synaptic phosphoprotein dysregulation (BIG2/KIF3A/GluN2A↓)
- Mitochondrial dynamics imbalance (DNM1L↓ & MFN1↑) → ATP↓40% + ROS↑250%
- Core Signaling Pathology:
- Sustained RASA2 overexpression → RAS/MAPK inhibition → synaptic impairment
- Neurodegenerative pathway activation: PI3K-AKT↑ & calcium signaling↑ (KEGG P<10⁻¹⁰)
- Therapeutic Discovery:
- Validated 10 target nodes (GPx-1, MAP1LC3B, PKCγ, etc.)
- Novel intervention: Functional restoration of RAS signaling → synaptic rescue
- Paradigm-Shifting Contribution:
- First dynamic molecular atlas of presymptomatic APOE4 pathology, establishing RASA2-RAS axis as preclinical Alzheimer's intervention target.
Signaling Pathway Analysis: Methodological Advancements and Clinical Translation
Innovative Framework:
- Tissue Sampling Rigor: Matched HCC tumor vs. histologically normal adjacent tissues (>1cm from capsule)
- Phosphoproteomic Workflow:
- TiO₂ magnetic bead phosphopeptide enrichment
- 6-plex TMT labeling + SCX fractionation (enhancing low-abundance detection)
- Q Exactive HF-MS (70,000 resolution) for deep profiling
Overcoming Traditional Limitations:
| Bottleneck | Current Breakthrough |
|---|---|
| Low-abundance signal loss | 5× sensitivity gain (858 differential phosphoproteins) |
| Early HCC marker deficiency | 14 core prognostic molecules identified (e.g., TOP2A/STMN1) |
Key Findings:
- Phosphorylation Landscape:
- 858 differentially phosphorylated proteins (1,326 phosphosites)
- 33 dysregulated pathways: spliceosome (P=1.3×10⁻⁸), HIF-1 (P=4.2×10⁻⁵), glycolytic reprogramming
- Clinical Validation:
- Prognostic markers: High MCM2/HJURP expression → 40% reduced median survival
- Therapeutic targeting: STMN1 phosphorylation correlates with sorafenib sensitivity (r=0.68, p<0.01)
- Novel target: ACOX inhibitors via fatty acid degradation pathway (P=0.003)
Translational Impact:
- Diagnostic: Early HCC phosphoscoring model (PNPLA7/ANLP-based)
- Therapeutic: TOP2A dephosphorylation strategy overcoming chemoresistance
- Precision Medicine: Integrated phosphoproteomics-transcriptomics (TCGA) PPPM framework
Conclusion
Phosphoproteomics represents a transformative analytical approach with demonstrated significance in fundamental biological research. While clinical translation encounters persistent technical and translational barriers, advancing methodologies and standardized protocols will increasingly position phosphoproteomic biomarkers as vital components in diagnostic frameworks and tailored therapeutic strategies. Looking forward, this discipline is poised to become integral to precision medicine paradigms, delivering critical molecular insights for early disease detection, therapeutic monitoring, and treatment efficacy assessment.
References
- Wu X, Liu YK, Iliuk AB, Tao WA. "Mass spectrometry-based phosphoproteomics in clinical applications." Trends Analyt Chem. 2023 Jun;163:117066. doi: 10.1016/j.trac.2023.117066
- Xu R, Chen Y, Wang Z, Zhang C, Dong X, Yan Y, Wang Y, Zeng Y, Chen P. "Phosphoproteomics Identifies Significant Biomarkers Associated with the Proliferation and Metastasis of Prostate Cancer." Toxins (Basel). 2021 Aug 9;13(8):554. doi: 10.3390/toxins13080554
- Martinez-Val A, Bekker-Jensen DB, Steigerwald S, Koenig C, Østergaard O, Mehta A, Tran T, Sikorski K, Torres-Vega E, Kwasniewicz E, Brynjólfsdóttir SH, Frankel LB, Kjøbsted R, Krogh N, Lundby A, Bekker-Jensen S, Lund-Johansen F, Olsen JV. "Spatial-proteomics reveals phospho-signaling dynamics at subcellular resolution." Nat Commun. 2021 Dec 7;12(1):7113. doi: 10.1038/s41467-021-27398-y
- Borek WE, Nobre L, Pedicona SF, Campbell AE, Christopher JA, Nawaz N, Perkins DN, Moreno-Cardoso P, Kelsall J, Ferguson HR, Patel B, Gallipoli P, Arruda A, Ambinder AJ, Thompson A, Williamson A, Ghiaur G, Minden MD, Gribben JG, Britton DJ, Cutillas PR, Dokal AD. "Phosphoproteomics predict response to midostaurin plus chemotherapy in independent cohorts of FLT3-mutated acute myeloid leukaemia." EBioMedicine. 2024 Oct;108:105316. doi: 10.1016/j.ebiom.2024.105316
- Lin YT, Way GP, Barwick BG, Mariano MC, Marcoulis M, Ferguson ID, Driessen C, Boise LH, Greene CS, Wiita AP. "Integrated phosphoproteomics and transcriptional classifiers reveal hidden RAS signaling dynamics in multiple myeloma." Blood Adv. 2019 Nov 12;3(21):3214-3227. doi: 10.1182/bloodadvances.2019000303
- Goetze S, van Drogen A, Albinus JB, Fort KL, Gandhi T, Robbiani D, Laforte V, Reiter L, Levesque MP, Xuan Y, Wollscheid B. "Simultaneous targeted and discovery-driven clinical proteotyping using hybrid-PRM/DIA." Clin Proteomics. 2024 Apr 2;21(1):26. doi: 10.1186/s12014-024-09478-5
- Wei P, Lin K, Chen X, Fang C, Qiu L, Hu J, Chang J. "Sequential Proteomic Analysis Reveals the Key APOE4-Induced Pathological and Molecular Features at the Presymptomatic Stage in Alzheimer's Disease Mice." CNS Neurosci Ther. 2025 Mar;31(3):e70306. doi: 10.1111/cns.70306
- Meng S, Li T, Wang T, Li D, Chen J, Li H, Cai W, Zeng Z, Liu D, Tang D, Hong X, Dai Y. "Global Phosphoproteomics Unveils Kinase-Regulated Networks in Systemic Lupus Erythematosus." Mol Cell Proteomics. 2022 Dec;21(12):100434. doi: 10.1016/j.mcpro.2022.100434
- Zhang Y, Li N, Yang L, Jia W, Li Z, Shao Q, Zhan X. "Quantitative phosphoproteomics reveals molecular pathway network alterations in human early-stage primary hepatic carcinomas: potential for 3P medical approach." EPMA J. 2023 Aug 10;14(3):477-502. doi: 10.1007/s13167-023-00335-3











