Peptide Sequencing Service
Creative Proteomics provides advanced peptide sequencing services to support research, drug development, and quality control. Our solutions cover de novo peptide sequencing, database-driven identification, and classical Edman degradation. We help clients identify unknown peptide sequences, confirm peptide integrity, and resolve sequencing problems caused by modifications or low-abundance peptides.
Problems We Solve
- Identify peptides with no reference database
- Confirm therapeutic peptide integrity for QC
- Analyse modifications and post-translational changes
- Resolve sequencing challenges in complex samples
Submit Your Request Now
×
- Overview
- Challenges
- Methods
- Workflow
- Platform
- Applications
- Sample Requirements
- Case Study
- FAQs
What is Peptide Sequencing?
Peptide sequencing is the process of determining the exact order of amino acids in a peptide chain. It plays a central role in proteomics, biomarker discovery, and therapeutic peptide development. Researchers rely on sequencing data to confirm peptide structures, verify synthetic products, and explore unknown bioactive peptides.
Two Core Strategies
Database Search Methods
Peptide sequencing by mass spectrometry often involves comparing fragmentation data against established protein sequence databases. This approach is efficient and reliable when a high-quality database is available. However, it becomes limited for novel peptides, non-model organisms, or modified structures.
De Novo Peptide Sequencing
When no database is available, de novo peptide sequencing reconstructs sequences directly from MS/MS fragment ions. By analysing the mass differences between b- and y-ions, researchers can infer amino acid residues step by step. This method solves common peptide sequencing problems, such as identifying new therapeutic peptides or sequencing peptides from uncharacterised species.
Other Peptide Sequencing Methods
- Peptide sequencing by Edman degradation remains useful for reading N-terminal residues, though its throughput is limited.
- Peptide sequencing using mass spectrometry with high-resolution Orbitrap or Q-TOF platforms delivers speed and accuracy for complex samples.
- Peptide sequencing by tandem mass spectrometry (MS/MS) is the most widely used approach today, especially for both database search and de novo sequencing.
- Nanopore peptide sequencing is an emerging method under investigation, offering potential for long-read peptide analysis.
By combining these approaches, Creative Proteomics ensures that even challenging peptides—such as those with modifications or low abundance—are accurately sequenced.
Why Peptide Sequencing Matters
Peptide sequencing is not just a technical exercise—it is a foundation for reliable discovery, drug development, and quality control. For research teams and pharmaceutical developers, sequencing data directly influences decision-making and regulatory outcomes.
Common Challenges in Peptide Research
- Database Gaps: Many bioactive peptides and therapeutic candidates are not represented in existing databases, making database-driven sequencing incomplete.
- Modified Structures: Post-translational modifications (PTMs) or chemical substitutions alter peptide mass, complicating routine analysis.
- Low Abundance: Rare or low-copy peptides are often undetectable with standard workflows.
- Regulatory Pressure: For peptide therapeutics, incomplete sequence confirmation can delay or block approval.
How Creative Proteomics Addresses These Issues
Our peptide sequencing service is designed to overcome these hurdles:
- De novo peptide sequencing reconstructs full sequences without relying on existing databases.
- Peptide sequencing by tandem mass spectrometry ensures accurate detection of modifications and rare peptides.
- Integrated workflows combine database search, de novo sequencing, and Edman degradation to maximise confidence.
- Regulatory-ready data provides clients with sequence confirmation suitable for QC, submission, and publication.
By solving these peptide sequencing problems, Creative Proteomics enables clients to validate novel peptides, accelerate therapeutic development, and meet compliance expectations.
Our Sequencing Methods
Creative Proteomics integrates multiple peptide sequencing methods to meet diverse research and development needs. Each approach is selected based on sample type, project goals, and data requirements.
Key Methods We Offer
Peptide Sequencing by Mass Spectrometry (MS/MS):
High-resolution Orbitrap and Q-TOF platforms deliver fast, accurate results for both simple and complex peptides.
De Novo Peptide Sequencing:
Essential for unknown peptides or when no database is available. Mass differences between fragment ions (b/y) are used to reconstruct amino acid order.
Database-Driven Sequencing:
Efficient when reference sequences exist, providing high-throughput peptide identification.
Peptide Sequencing by Edman Degradation:
A classical N-terminal sequencing technique, often applied to shorter peptides or confirmation studies.
Nanopore Peptide Sequencing:
An emerging method that enables continuous readout of amino acids, currently used in exploratory research.
Comparison of Peptide Sequencing Methods
| Method | Strengths | Limitations | Typical Applications |
|---|---|---|---|
| De Novo Peptide Sequencing | Independent of databases; resolves unknown peptides; detects modifications | Computationally intensive; may require high-quality spectra | Novel peptide discovery, rare species, synthetic peptides |
| Database-Driven Sequencing (MS/MS Search) | Fast, reliable when references exist; scalable for large datasets | Limited to known sequences in databases | Routine proteomics, QC with established references |
| Peptide Sequencing by Edman Degradation | Accurate N-terminal readout; complementary to MS/MS | Low throughput; ineffective for long/blocked peptides | N-terminal validation, short synthetic peptides |
| Peptide Sequencing by Tandem MS/MS | High sensitivity; robust for modifications; industry-standard | Requires fragmentation quality and skilled interpretation | Therapeutic validation, biomarker discovery |
| Nanopore Peptide Sequencing (Emerging) | Potential for long-read peptide sequencing; single-molecule analysis | Limited availability; still under development | Exploratory proteomics, next-generation R&D |
By combining de novo, Edman, and MS/MS-based methods, our platform ensures accurate results even for highly complex or modified peptides.
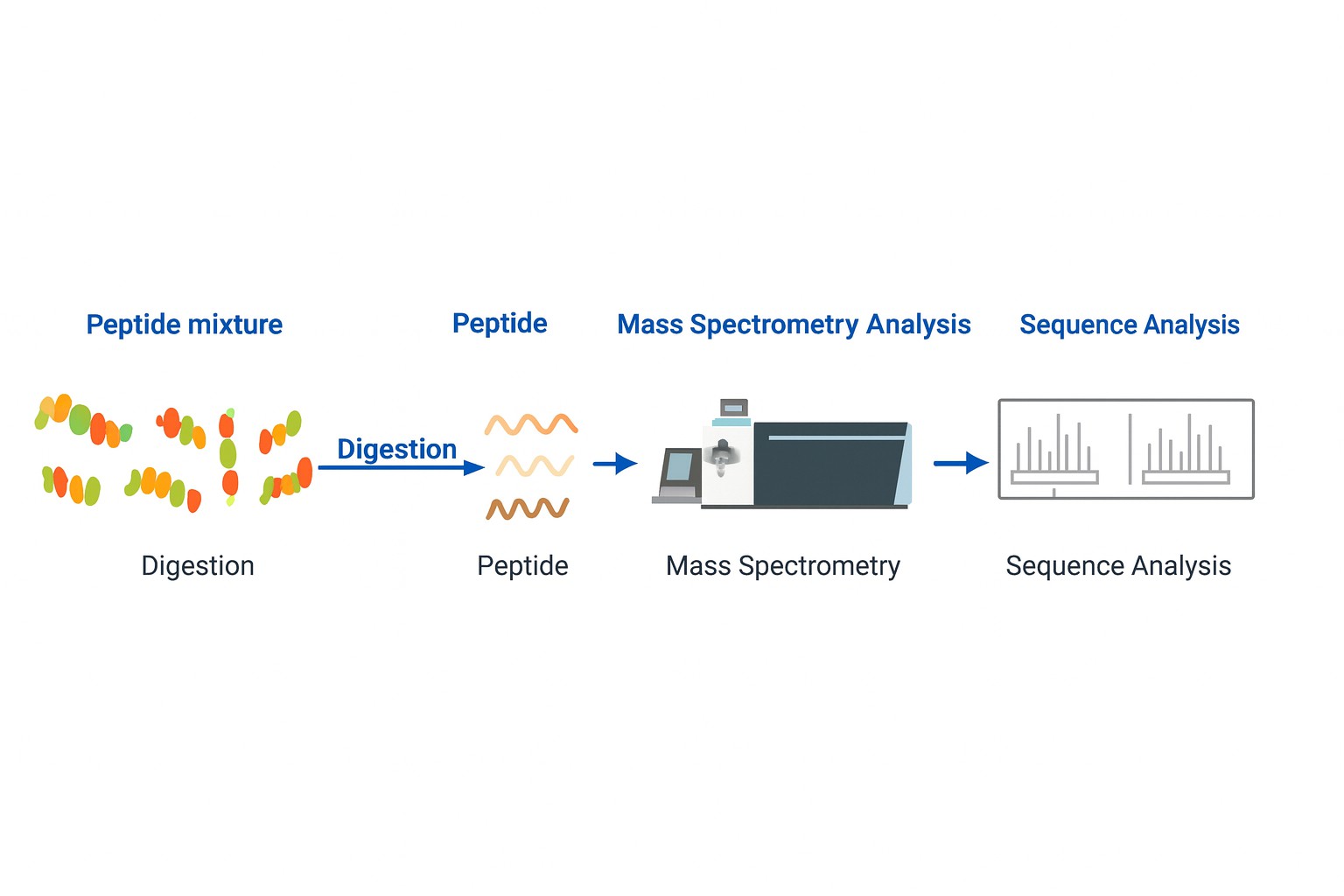
Workflow
Our peptide sequencing workflow combines advanced mass spectrometry methods with rigorous bioinformatics analysis to ensure accurate, reproducible results. Each stage is optimised for sensitivity, reliability, and data clarity.
Step 1. Sample Preparation
Clients provide purified peptides in lyophilised or solution form. We recommend ≥20 µg per sample, free from high salts or detergents. Proper preparation improves fragmentation quality and ensures consistent sequencing outcomes.
Step 2. Enzymatic Digestion and MS/MS Analysis
Samples are digested with one or more proteases to generate overlapping fragments. These fragments are analysed by tandem mass spectrometry (MS/MS) on high-resolution platforms such as Orbitrap Fusion Lumos or Q-TOF.
Step 3. Fragment Ion Mapping
Collision-induced dissociation (CID), higher-energy collisional dissociation (HCD), and electron transfer dissociation (ETD) generate fragment ions (b/y, a/x). Mass differences between these ions provide the basis for amino acid sequence reconstruction.
Step 4. De Novo Assembly and Database Search
- De novo sequencing algorithms reconstruct sequences directly from fragment spectra.
- Database search tools validate results against existing protein or peptide libraries.
- Combined approaches maximise accuracy and reduce ambiguity, especially for modified or unknown peptides.
Step 5. Data Verification and Reporting
Bioinformatics pipelines integrate sequencing results, coverage maps, and modification detection. Final deliverables include FASTA sequences, annotated spectra, and publication-ready figures. Expert review ensures each sequence is fully validated.
Our Technology Platform
Creative Proteomics operates on an advanced mass spectrometry–based peptide sequencing platform designed for accuracy, sensitivity, and flexibility.
Orbitrap Fusion Lumos Mass Spectrometer
Provides ultra-high resolution and mass accuracy, enabling reliable sequencing of peptides with modifications or low abundance. The instrument supports multiple fragmentation techniques (CID, HCD, ETD) for complete peptide coverage.

Nano-LC (Nanoliter Chromatography) System
Enhances separation efficiency before MS detection, ensuring clean spectra even in complex mixtures. This is essential for de novo peptide sequencing and post-translational modification analysis.

Applications of Peptide Sequencing
Peptide sequencing provides actionable insights across life science research, therapeutic development, and product quality control. By delivering precise amino acid sequence information, it supports multiple scientific and industrial workflows.
Drug Discovery and Development
- Confirm the identity of synthetic peptide drugs before preclinical or clinical studies.
- Detect sequence variants or modifications that may affect therapeutic efficacy.
- Support the discovery of cytokine peptide mimetics, small peptides that mimic cytokine activity but have distinct sequences and lower molecular weight. Many of these candidates are currently in preclinical or clinical testing.
- Provide regulatory-compliant data packages for peptide-based submissions.
Biomarker Discovery and Diagnostics
- Identify novel peptide biomarkers from biological fluids or tissues.
- Support diagnostic research by sequencing disease-related peptides.
- Enable validation of peptide biomarkers for precision medicine initiatives.
- Develop diagnostic peptide antigens, which are more specific and easier to prepare than natural proteins. These are widely used in test kits for viruses (HIV, HBV, HCV), bacteria, parasites, and autoimmune markers.
Protein Engineering and Functional Studies
- Verify expression products in protein engineering and synthetic biology projects.
- Characterise structural variants introduced by mutagenesis or chemical modification.
- Provide sequence-level insights for downstream assays such as binding affinity studies.
- Support research on antibacterial active peptides, many of which show dual activity against pathogens and tumor cells. More than 100 such peptides have been reported and validated in vitro and in vivo.
Quality Control and Pharmaceutical Development
- Ensure batch-to-batch consistency of peptide APIs or intermediates.
- Detect impurities or degradation products during manufacturing.
- Sequence therapeutic small peptides for regenerative and anti-inflammatory medicine. Published studies show peptides that promote wound healing, reduce corneal inflammation, and inhibit bone resorption.
- Comply with guidelines such as USP 1503 Quality Attributes of Synthetic Peptide Drug Substances.
By addressing these areas, Creative Proteomics' peptide sequencing service strengthens decision-making in both academic and industrial projects.
Sample Requirements for Peptide Sequencing
| Sample Type | Recommended Quantity | Minimum Quantity | Notes |
|---|---|---|---|
| Purified Peptides (Lyophilised or Solution) | ≥20 µg | 10 µg | Avoid SDS, detergents, or high salt; store at –80°C |
| Protein Samples (for digestion & sequencing) | ≥50 µg | 20 µg | Provide in buffer without interfering chemicals; snap-freeze in liquid nitrogen |
| Cell Lysates (for peptide preparation) | ≥1 × 10⁶ cells | 5 × 10⁵ cells | Wash with PBS, avoid serum contamination, quick-freeze |
| Tissue Samples | ≥50 mg | 20 mg | Fresh-frozen in liquid nitrogen; divide into aliquots to avoid repeated freeze–thaw |
| Plasma/Serum | ≥1 mL | 500 µL | Centrifuge to remove cells/debris; freeze immediately at –80°C |
General Instructions
- Use 1.5 mL centrifuge tubes with screw caps; seal with parafilm for shipment.
- Label tubes clearly with sample ID matching the submission form.
- Transport solid samples on ice packs and liquid samples on dry ice.
- Submit an electronic copy of the completed sample submission form along with QC data.
Client Case Study: Proteomic Sequencing of Splenic Changes Following Neural Intervention

- Background
- Methods
- Results
- Conclusions
A research group investigated how adolescent brain development influences peripheral immune function. Specifically, they studied the effect of inhibiting microglial synaptic pruning in the nucleus accumbens (NAc) on splenic protein expression in rats. To uncover global proteomic changes, the team partnered with Creative Proteomics for high-resolution peptide and protein sequencing support.
Spleen samples were collected from male and female rats subjected to NAc intervention. Creative Proteomics performed label-free nanoLC-MS/MS proteomic sequencing:
- Protein extraction and trypsin digestion
- LC-MS/MS analysis on high-resolution Orbitrap systems
- De novo peptide sequencing combined with database-driven identification
- Bioinformatics pipelines for differential protein expression analysis
This integrative approach ensured accurate detection of low-abundance peptides and post-translational modifications, which are common challenges in peptide sequencing by mass spectrometry.
Creative Proteomics identified nearly 3,000 proteins per condition, with over 290 differentially expressed proteins (DEPs) in both male and female spleens. Importantly, the sequencing data revealed:
- A high degree of overlap in global proteomic changes across sexes
- One male-specific downregulated protein (Phf11l), linked to IL-2 regulation
- Six female-specific DEPs associated with neurochemical and immune functions
The client used these results to validate downstream cytokine assays, confirming sex-specific immune modulation.
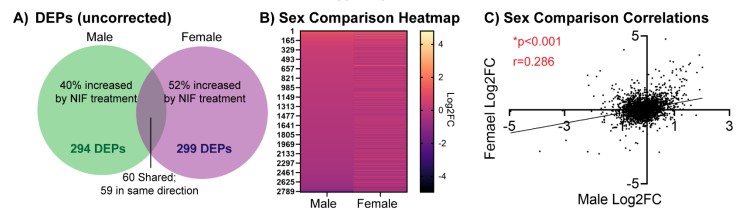 Figure 1. Global proteomic changes identified by Creative Proteomics' peptide sequencing service. Inhibition of microglial pruning in the NAc altered splenic protein expression, with 2930 proteins identified in males and 2933 in females, and significant overlaps between sexes.
Figure 1. Global proteomic changes identified by Creative Proteomics' peptide sequencing service. Inhibition of microglial pruning in the NAc altered splenic protein expression, with 2930 proteins identified in males and 2933 in females, and significant overlaps between sexes.
This project highlights how Creative Proteomics' peptide sequencing by tandem mass spectrometry enables researchers to uncover complex, sex-specific proteomic responses. By delivering comprehensive, publication-ready sequencing data, Creative Proteomics allowed the client to:
- Resolve sequencing problems caused by low-abundance proteins
- Identify regulatory proteins linked to cytokine expression
- Provide mechanistic insights supporting peer-reviewed publication
Reference
- Kirkland JM, Patel I, Ardeshna MS, Kopec AM. Microglial synaptic pruning in the nucleus accumbens during adolescence sex-specifically influences splenic immune outcomes. bioRxiv. 2023; doi: 10.1101/2023.05.03.539317. Available under a CC-BY 4.0 International license.
Demo for Protein Mass Spectrometry Service
Figures come from (Shibata, H.et.al, J Clin Immunol,2024)
Publications

- KUMAR, L. K., Han, J., Dalvi, S., Foley, N., Subedi, Y., & Singh, R. Impaired phagocytosis of photoreceptor outer segments by RPE in CLN3 disease is a consequence of altered sphingolipid metabolism. Investigative Ophthalmology & Visual Science
- Kirkland, J. M., Patel, I., & Kopec, A. M. Microglia-mediated synaptic pruning in the nucleus accumbens during adolescence: A preliminary study of the proteomic consequences and putative female-specific pruning target. bioRxiv
Frequently Asked Questions (FAQs)
What is de novo peptide sequencing and when should I use it?
De novo peptide sequencing is a method where the amino acid sequence is determined directly from MS/MS fragment ions, without relying on any protein database. It is ideal when you have novel peptides, unusual modifications, or samples from organisms with incomplete sequence databases. Using this method avoids "peptide sequencing by database search" limitations.
How does peptide sequencing by mass spectrometry compare to Edman degradation?
Mass spectrometry (especially LC-MS/MS) is more versatile and sensitive than Edman degradation. It handles modified residues, low-abundance peptides, and complex samples better. Edman degradation is useful for confirming N-terminal residues in simpler or purified peptides, but is less effective when the peptide is blocked at the N-terminus or contains unusual amino acids.
Can your service resolve peptide sequencing problems caused by modifications or low signal?
Yes. We use high-resolution mass spectrometers combined with multiple fragmentation methods (e.g. CID, HCD, ETD) plus de novo sequencing algorithms. This combination increases detection sensitivity for modified peptides or fragments with weak signal. Also, our expert review helps assign ambiguous residues.
Do I need a database for peptide sequencing?
Not always. If the peptide or protein sequence is known and represented in existing databases, database-driven MS/MS search works well. But for unknown peptides, novel modifications, or for organisms without well-annotated databases, de novo sequencing lets us reconstruct the full sequence from scratch.
What sample quality or preparation is required for accurate peptide sequencing?
A purified peptide sample helps—buffers should be free of high salt, detergent, or SDS. Solid (lyophilised) peptides or clean solution forms work well. Sufficient peptide quantity and avoiding contaminants improve fragmentation quality and overall confidence in sequence assignments.
What kinds of deliverables can I expect from your peptide sequencing service?
You will receive validated amino acid sequences (e.g., FASTA format), annotated MS/MS spectra, coverage maps showing which peptide fragments support each residue, a report of detected modifications, and publication-quality visuals.
What are the limitations of peptide sequencing by tandem mass spectrometry?
Some limitations include: ambiguous assignment of isoleucine vs leucine (they have the same mass), blocked N-termini preventing Edman degradation, very low abundance peptides that fall below detection limit, and extreme modifications that significantly disrupt fragmentation patterns. Proper sample prep and using multiple fragmentation modes help mitigate these issues.
Learn about other Q&A.
References
- Keough T, Youngquist RS, Lacey MP. A method for high-sensitivity peptide sequencing using postsource decay matrix-assisted laser desorption ionization mass spectrometry. Proc Natl Acad Sci U S A. 1999 Jun 22;96(13):7131-6. doi: 10.1073/pnas.96.13.7131. PMID: 10377380; PMCID: PMC22028.
- Liu, K., Ye, Y., Li, S. et al. Accurate de novo peptide sequencing using fully convolutional neural networks. Nat Commun 14, 7974 (2023).
- Xiao-Mei Sha, Le-Ying Yang, Hui Wang, Jie Cheng, Hao-Sheng Luo, Li-Jun Zhang, Zi-Zi Hu, Zong-Cai Tu, Identifying gelatin source by mass spectrometry: A review, Food Chemistry, 10.1016/j.foodchem.2025.144904, 488, (144904), (2025).




















