Omega-3 Fatty Acids Analysis Service
Creative Proteomics provides accurate and comprehensive omega-3 fatty acid analysis using advanced techniques like LC-MS/MS and GC-MS. Our services include quantification, lipid profiling, isomer analysis, and metabolic pathway mapping, helping companies in the food, nutraceutical, and cosmetic industries optimize product formulations, ensure quality control, and support research and development. By offering tailored solutions, we address the need for precise omega-3 content analysis, improving product integrity and advancing health-focused innovations.
Submit Your Request Now
×- What We Provide
- Advantage
- Workflow
- Technology Platform
- Sample Requirements
- FAQ
- Publication
What is Omega-3 Fatty Acids?
Omega-3 fatty acids are a class of essential polyunsaturated fatty acids (PUFAs) characterized by the presence of a double bond at the third carbon atom from the methyl end of the fatty acid chain. These molecules are widely recognized for their structural and functional roles in biological membranes, as well as their significance in nutritional formulations and industrial lipid applications. The most commonly studied omega-3 fatty acids include:
- Alpha-linolenic acid (ALA; C18:3n-3) – a plant-derived essential fatty acid
- Eicosapentaenoic acid (EPA; C20:5n-3) – prevalent in marine oils
- Docosahexaenoic acid (DHA; C22:6n-3) – important in cellular lipid composition
- Docosapentaenoic acid (DPA; C22:5n-3) – an intermediate in metabolic conversion
These compounds are not synthesized efficiently by humans or animals and must be obtained through dietary sources such as fish oil, algal oil, flaxseed, and fortified foods. They are widely monitored and optimized in product formulations across the functional food, nutraceutical, agriculture, and cosmetic sectors.
Why Analyze Omega-3 Fatty Acids?
Analyzing omega-3 fatty acids is crucial for product development, quality assurance, and research purposes. Accurate analysis supports various applications:
- Formulation optimization: Ensures balanced omega-3 content in food, nutraceutical, and cosmetic products.
- Quality control: Confirms the consistency and purity of omega-3 in supplements and other products.
- Research and development: Facilitates the study of omega-3's role in metabolic pathways and its impact on health.
- Process optimization: Enhances extraction methods and refines production processes for raw materials.
Omega-3 analysis ensures product integrity, supports product development, and offers valuable insights for further research.
Omega-3 Fatty Acids Analysis Service Offered by Creative Proteomics
Quantification of Omega-3 Fatty Acids
- EPA, DHA, ALA, DPA, and other omega-3s are accurately quantified using LC-MS/MS and GC-MS techniques.
- Absolute quantification with internal standards ensures precise and reproducible results across multiple sample types.
Comprehensive Lipid Profiling
- Full lipidomic analysis including omega-3 fatty acids and their derivatives.
- Detection of key omega-6/omega-3 ratios to assess nutritional balance.
- Metabolites like resolvins and protectins are also identified to explore functional effects.
Isomer Profiling and Differentiation
- Analysis of DHA/EPA isomers and differentiation of omega-3 variants.
- Detection of Saturated and unsaturated isomers for in-depth product characterization.
Metabolomic Pathway Analysis
- Pathway mapping of omega-3 fatty acids and their metabolites through integrated bioinformatics tools.
- Identification of key pathways involved in lipid metabolism and inflammation resolution.
Matrix-Specific Extraction and Analysis
Customized sample extraction methods for a wide variety of matrices, including:
- Fish oils, algae oils, and plant-based sources
- Plasma, serum, tissues, and other biological samples
- Nutritional products and fortified foods
- Fermentation and production-derived samples
Stability and Shelf-Life Testing
Oxidative stability testing of omega-3-rich products. Long-term storage and shelf-life studies to monitor degradation and quality control.
Custom Analytical Services
Development of custom lipidomic panels tailored to specific research or product needs. Targeted analysis of additional fatty acids and related metabolites upon request.
High-Throughput Screening
Support for high-throughput analysis, enabling rapid screening of large sample sets. Automated sample preparation and LC-MS/MS workflows provide quick and reliable results.
List of Detected Omega-3 Fatty Acids and Related Metabolites
| Category | Compounds Detected | Related Metabolic Pathways |
|---|---|---|
| Omega-3 Fatty Acids | Alpha-linolenic acid (ALA, C18:3n-3) | Lipid metabolism, inflammation resolution |
| Eicosapentaenoic acid (EPA, C20:5n-3) | Omega-3 to EPA conversion, anti-inflammatory pathways | |
| Docosahexaenoic acid (DHA, C22:6n-3) | Neurogenesis, synaptic plasticity, anti-inflammatory pathways | |
| Docosapentaenoic acid (DPA, C22:5n-3) | Omega-3 metabolic conversion to EPA and DHA | |
| Omega-3 Metabolites | Resolvin D1, D2 (from EPA) | Resolution of inflammation, lipid mediator signaling |
| Protectin D1 (from DHA) | Anti-inflammatory, neuroprotective properties | |
| Maresin 1 (from DHA) | Inflammation resolution, tissue repair | |
| 14-Hydroxy-docosahexaenoic acid (14-HDHA) | Neuroprotective pathways, oxidative stress response | |
| Oxidized Lipids | Hydroxyeicosapentaenoic acid (HEPE) | Oxidative stress, cellular signaling |
| Epoxyeicosatetraenoic acid (EET) | Vascular function, inflammation regulation | |
| Omega-6/Omega-3 Ratios | Arachidonic acid (ARA, C20:4n-6) | Pro-inflammatory signaling, eicosanoid production |
| Linoleic acid (LA, C18:2n-6) | Omega-6 pathway, balance with Omega-3 | |
| Beta-Oxidation Intermediates | Acyl-carnitines (e.g., C20:5-carnitine, C22:6-carnitine) | Mitochondrial lipid catabolism and energy metabolism |
Advantages of Omega-3 Fatty Acids Assay
- Ultra-high sensitivity, with detection limits as low as 0.5 ng/mL, enables trace-level quantification of EPA, DHA, and related metabolites in complex matrices.
- Extensive compound coverage, detecting over 50 omega-3-related fatty acids and metabolic intermediates across multiple biosynthetic pathways.
- Exceptional reproducibility, with intra- and inter-batch coefficient of variation (CV) consistently maintained below 10% using isotope-labeled internal standards.
- Accelerated turnaround time, delivering results within 14–42 business days while supporting high-throughput capacity of up to 200 samples per day.
- Scalable analytical platform, suitable for both small-batch product evaluation and large-cohort research studies, supported by automated sample preparation and LC-MS workflows.
- Matrix versatility, accommodating a wide range of sample types including fish oils, microalgae, fortified foods, plasma, serum, tissues, and fermentation broths.
- Robust quantification range, from 0.5 ng/mL to over 100 µg/mL, meeting the needs of both trace detection and bulk compositional analysis.
- Comprehensive pathway interpretation, with detected metabolites mapped to KEGG, HMDB, and LIPID MAPS databases for full lipid pathway contextualization.
- Customized reporting formats, including concentration tables, metabolic pathway visualizations, and statistical summaries tailored to client-specific research or development objectives.
Workflow for Omega-3 Fatty Acids Analysis Service
Technology Platform for Omega-3 Fatty Acids Analysis Service
LC-MS/MS (Thermo Scientific Q Exactive™ Orbitrap) – For high-resolution quantification.
Agilent 6545XT AdvanceBio Q-TOF – For targeted lipidomics and fragmentation profiling.
GC-MS (Agilent 7890B-5977B) – For volatile and derivatized fatty acids.
NMR Spectroscopy (Bruker Avance III 600 MHz) – For absolute structural confirmation.

Thermo Fisher Q Exactive (Figure from Thermo Fisher)

Agilent 7890B-5977B (Figure from Agilent)

Bruker Avance III 600 MHz (Fig from Bruker)
Sample Requirements for Omega-3 Fatty Acids Analysis Service
| Sample Type | Required Amount | Notes |
|---|---|---|
| Plasma / Serum | ≥ 200 µL | Use EDTA or heparin tubes; avoid hemolysis |
| Whole Blood | ≥ 500 µL | Store at −80°C; freeze immediately after collection |
| Tissue (e.g., liver, brain) | ≥ 100 mg | Snap-frozen in liquid nitrogen; store at −80°C |
| Cell Pellets | ≥ 1 × 10⁷ cells | Washed with PBS; flash frozen |
| Fish Oil / Algae Oil | ≥ 100 µL | Store in amber vials; protect from light and air |
| Food or Feed Products | ≥ 1 g | Homogenize thoroughly; store at −20°C or lower |
| Fermentation Broth / Culture Supernatant | ≥ 2 mL | Filtered or centrifuged to remove particulates |
| Plant Material / Seeds | ≥ 100 mg (dry weight) | Freeze-dried and ground preferred |
| Supplement Capsules | ≥ 2 capsules or softgels | Submit unopened or extracted oil in sealed vials |
Omega-3 Fatty Acids Analysis Service Case Study

Title: Omega-3 Polyunsaturated Fatty Acids Decrease Aortic Valve Disease Through the Resolvin E1 and ChemR23 Axis
Journal: Circulation
Published: 2020
- Background
- Materials & Methods
- Results
- Reference
Aortic valve stenosis (AVS) is the most common valvular heart disease, characterized by progressive narrowing of the aortic valve due to leaflet thickening and calcification. It significantly increases the risk of heart failure and cardiovascular mortality. Traditional treatments fail to halt disease progression, leaving valve replacement as the only option. Omega-3 polyunsaturated fatty acids (n-3 PUFAs), particularly EPA and DHA, have demonstrated cardiovascular benefits due to their anti-inflammatory and proresolving properties, which are mediated through specialized proresolving mediators (SPMs) like resolvin E1 (RvE1). This study investigates the role of RvE1 and its receptor ChemR23 in reducing AVS progression.
The study analyzed tricuspid aortic valves from 96 patients undergoing valve replacement surgery, with full ethical approval and informed consent. Lipidomic profiling was performed using liquid chromatography–tandem mass spectrometry to quantify specialized proresolving mediators. For mechanistic studies, genetically modified mice—including Fat-1 transgenic, Apoe−/−, ChemR23−/−, and their combinations—were used. Statistical analyses involved t-tests, ANOVAs, and mixed-effects models, with significance set at P<0.05. All procedures followed ethical standards for human and animal research.
n-3 PUFA Content in Human Aortic Valves
Noncalcified aortic valve tissue had significantly higher HS-omega-3 index compared to calcified regions. Slower AVS progression was associated with a higher HS-omega-3 index, though not statistically significant.
Lipid Mediators in Aortic Valves
n-3 PUFA-derived SPMs (RvE1 and RvD3) were lower in calcified valves. In contrast, the pro-inflammatory leukotriene B4 from n-6 PUFA was higher in calcified tissue.
RvE1 Receptor (ChemR23) in Aortic Valves
ChemR23, the RvE1 receptor, was predominantly expressed in human aortic valves and was associated with reduced calcification in vitro.
Fat-1tg×Apoe−/− Mice
These mice, with endogenous n-3 PUFA production, showed lower transaortic velocity, increased cusp separation, and reduced valve calcification compared to controls.
ChemR23 as a Key Mediator
ChemR23 deletion in Apoe−/− mice reversed the benefits of n-3 PUFAs, leading to increased Vmax, reduced cusp separation, and increased leaflet calcification.
Inflammation and Macrophage Polarization
Fat-1tg promoted M2 macrophage polarization (CD206, arginase-1) in a ChemR23-dependent manner, with no significant change in M1 markers.
Human Data Correlation
ChemR23 expression in human valves correlated with M2 macrophage markers, particularly in both calcified and noncalcified regions.
ChemR23 Deletion in Valve Injury Model
ChemR23−/− mice exhibited worse outcomes after aortic valve injury, including increased leaflet thickening and higher Vmax.
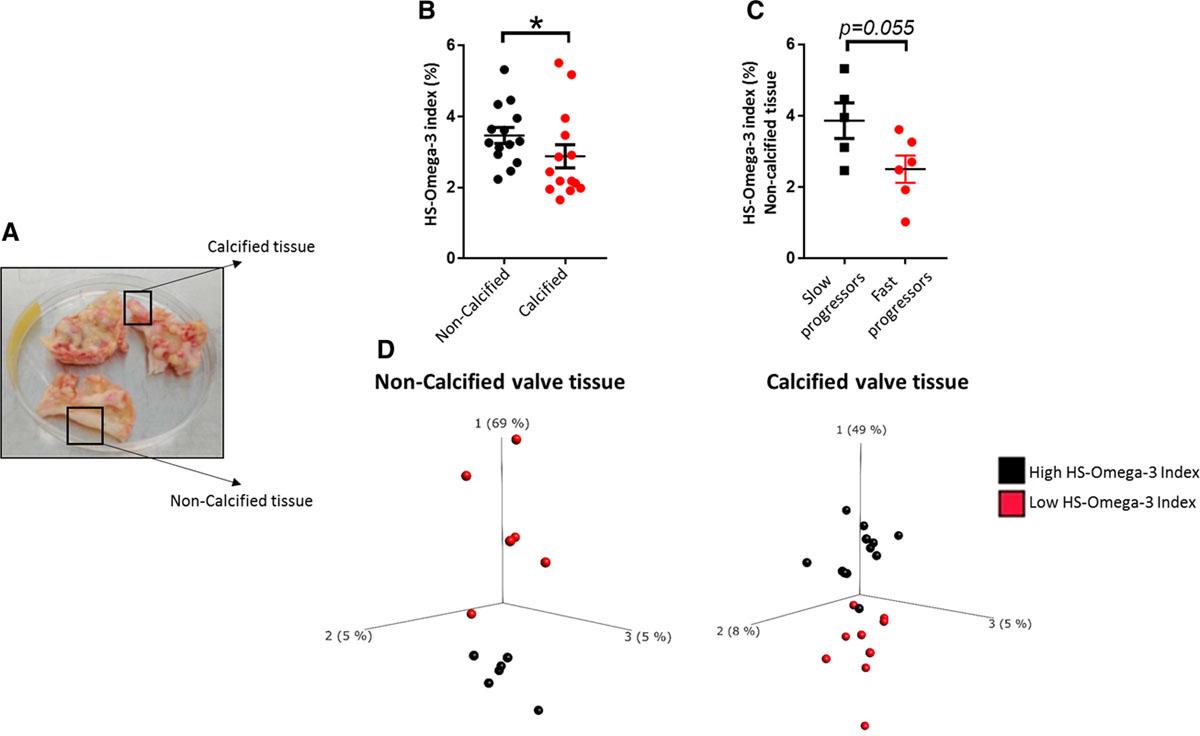 HS-omega-3 index is decreased in calcified aortic valve tissue and is lower in noncalcified valve tissue of aortic valve stenosis (AVS) fast progressors.
HS-omega-3 index is decreased in calcified aortic valve tissue and is lower in noncalcified valve tissue of aortic valve stenosis (AVS) fast progressors.
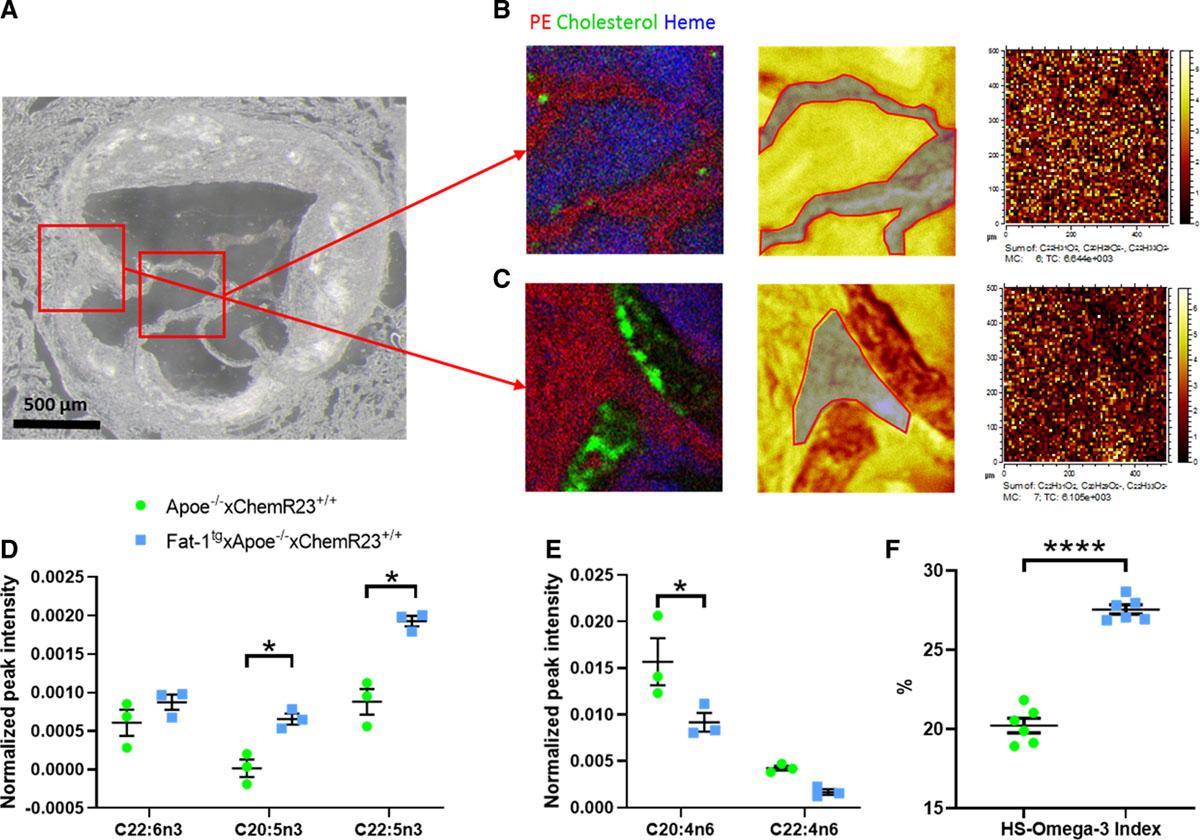 Omega-3 polyunsaturated fatty acids (PUFAs) are higher and n-6 PUFAs are lower in valve leaflets of Fat-1tg×Apoe−/− compared with Apoe−/− mice.
Omega-3 polyunsaturated fatty acids (PUFAs) are higher and n-6 PUFAs are lower in valve leaflets of Fat-1tg×Apoe−/− compared with Apoe−/− mice.
Reference
- Artiach, Gonzalo, et al. "Omega-3 polyunsaturated fatty acids decrease aortic valve disease through the resolvin E1 and ChemR23 axis." Circulation 142.8 (2020): 776-789. https://doi.org/10.1161/CIRCULATIONAHA.119.041868.
FAQ of Omega-3 Fatty Acids Analysis Service
Can omega-3 analysis differentiate between marine-based (fish oil) and plant-based (flaxseed) sources?
Yes. Our GC-MS methods distinguish between short-chain ALA (plant-derived, e.g., flaxseed) and long-chain EPA/DHA (marine-derived, e.g., fish oil) through precise retention time matching and fragmentation patterns. For example, ALA (C18:3n3) elutes earlier than EPA (C20:5n3) and DHA (C22:6n3), enabling clear source attribution.
How does sample storage time affect omega-3 stability?
Omega-3 fatty acids, particularly EPA and DHA, are prone to oxidation. Samples stored at -80°C retain stability for up to 12 months; repeated freeze-thaw cycles (>3 cycles) may degrade PUFAs by 10–15%, necessitating single-use aliquots .
What is the clinical significance of the omega-6/omega-3 ratio in analysis reports?
A high omega-6/omega-3 ratio (>10:1) is linked to pro-inflammatory states. Our service calculates this ratio using arachidonic acid (AA, C20:4n6) versus EPA+DHA levels, providing actionable insights for dietary adjustments to achieve optimal ratios (4:1 or lower) .
Can analysis detect oxidized omega-3 metabolites in degraded samples?
Yes. We employ GC-MS with derivatization to stabilize and quantify oxidation byproducts (e.g., malondialdehyde) in rancid oils or improperly stored samples, ensuring data accuracy even in compromised matrices.
How do omega-3 levels in breast milk correlate with maternal supplementation?
Maternal DHA intake directly impacts breast milk concentrations. Our analysis supports lactation studies by quantifying DHA levels, with typical ranges of 0.1–1.0% of total fatty acids. Supplementation (300–600 mg/day DHA) can increase milk DHA by 30–50% .
Are there interference risks from concurrent vitamin E (antioxidant) supplementation?
No. Vitamin E does not interfere with omega-3 quantification via GC-MS, as it elutes in a separate chromatographic region. However, high-dose antioxidants may mask oxidative degradation in samples, which we flag through peroxide value assessments.
Can your service analyze omega-3s in complex matrices like infant formula or fortified snacks?
Yes. We validate methods for lipid extraction from emulsified or high-carbohydrate matrices, achieving recovery rates >95% for EPA/DHA in infant formula and 85–90% in baked goods, despite challenges from encapsulation .
How does heat processing (e.g., cooking) affect omega-3 levels in food samples?
Grilling or frying reduces EPA/DHA by 15–30% due to thermal oxidation. Our analysis includes stability testing under controlled heating to simulate food processing effects.
Learn about other Q&A.
Publications
Here are some publications in Lipidomics research from our clients:

- Evidence for phosphate-dependent control of symbiont cell division in the model anemone Exaiptasia diaphana. 2024. https://doi.org/10.1128/mbio.01059-24
- Loss of G0/G1 switch gene 2 (G0S2) promotes disease progression and drug resistance in chronic myeloid leukaemia (CML) by disrupting glycerophospholipid metabolism. 2022. https://doi.org/10.1002/ctm2.1146
- Summative and ultimate analysis of live leaves from southern US forest plants for use in fire modeling. 2020. https://doi.org/10.1152/ajpgi.00184.2023
- Characterization of Dnajc12 knockout mice, a model of hypodopaminergia. 2024. https://doi.org/10.1101/2024.07.06.602343
- Multi‐omics identify xanthine as a pro‐survival metabolite for nematodes with mitochondrial dysfunction. 2019. https://doi.org/10.15252/embj.201899558









