Glycosaminoglycans Analysis Service
Creative Proteomics offers high-precision glycosaminoglycans (GAGs) analysis services using advanced UPLC-MS/MS and Orbitrap platforms to support basic research and biopharmaceutical development. Our service enables accurate quantification, structural characterization, and metabolic profiling of GAGs and related compounds, helping researchers unravel GAG functions, investigate drug mechanisms, and optimize bioprocesses with reliable, reproducible data.
Submit Your Request Now
×- What We Provide
- Advantage
- Workflow
- Technology Platforms
- Sample Requirements
- FAQ
- Publication
What are Glycosaminoglycans?
Glycosaminoglycans (GAGs) are a diverse group of highly sulfated, linear polysaccharides composed of repeating disaccharide units. They are key components of the extracellular matrix and are covalently linked to core proteins to form proteoglycans. GAGs play crucial roles in modulating cell signaling, maintaining tissue hydration, regulating enzymatic activity, and mediating interactions with growth factors, cytokines, and structural proteins. The major classes include heparan sulfate (HS), chondroitin sulfate (CS), dermatan sulfate (DS), keratan sulfate (KS), and hyaluronic acid (HA), each exhibiting distinct structural motifs and biological functions. Due to their structural complexity and biological relevance, comprehensive analysis of GAGs is essential for understanding cellular microenvironments and for advancing drug development, biomaterials research, and systems biology.
Glycosaminoglycans Analysis Service Offered by Creative Proteomics
- Total GAG Quantification: Quantification of total glycosaminoglycan content in biological samples (plasma, serum, tissues).
- Individual GAG Class Profiling: Identification and quantification of Heparan Sulfate (HS), Chondroitin Sulfate (CS), Dermatan Sulfate (DS), Keratan Sulfate (KS), and Hyaluronic Acid (HA).
- Chain Length Distribution Analysis: Characterization of GAG oligosaccharide chain lengths.
- Sulfation Pattern & Structural Elucidation: Analysis of sulfation patterns on GAG molecules.
- GAG Metabolite Profiling: Detection and quantification of GAG-related metabolites (e.g., UDP-sugars, sulfated intermediates).
- Pathway Mapping of GAG Biosynthesis & Degradation: Mapping GAG metabolic pathways using LC-MS/MS and computational models.
- Isoform Detection & Analysis: Detection and differentiation of GAG isoforms. Method: High-resolution MS/MS, isomer-specific separation.
- High Throughput GAG Screening: Large-scale screening of biological samples for GAG content.
- Custom GAGs Analysis Projects: Tailored GAG analysis services based on specific research needs.
List of Detected Glycosaminoglycans and Related Metabolites
| GAG Class | Detected Analytes |
|---|---|
| Heparan Sulfate (HS) | HS0S, HS2S, HS6S, HSNS, HS2S6S, HSNS2S, HSNS6S, HSTriS, Heparin, Heparan sulfate oligosaccharides |
| Chondroitin Sulfate (CS) | CS0S, CS4S, CS6S, CS2S4S, CS2S6S, CS4S6S, Chondroitin disaccharides |
| Dermatan Sulfate (DS) | DS0S, DS2S, DS4S, DS6S, DS2S4S, Dermatan sulfate oligosaccharides |
| Keratan Sulfate (KS) | KS-I, KS-II, Keratan sulfate disaccharides, Keratan sulfate oligosaccharides |
| Hyaluronic Acid (HA) | HA mono-, di-, tri-, and tetra-saccharides, Hyaluronan oligomers |
| Metabolites | Detected Compounds |
|---|---|
| UDP-Sugars | UDP-glucuronic acid, UDP-N-acetylglucosamine, UDP-N-acetylgalactosamine, UDP-glucose, UDP-galactose |
| Sulfated Intermediates | PAPS (3'-phosphoadenosine-5'-phosphosulfate), 3'-phosphoadenosine-5'-sulfate (PAPS), Sulfated sugars (e.g., N-acetylglucosamine-6-sulfate, galactose-6-sulfate) |
| Pathway-Linked Compounds | Detected Compounds |
|---|---|
| Metabolic Intermediates | S-adenosylmethionine (SAM), S-adenosylhomocysteine (SAH), ATP, NAD+, NADH, Coenzyme A |
Advantages of Glycosaminoglycans Assay
- High Sensitivity: Detection limits down to 0.1 ng/mL for sulfated GAGs using UPLC-MS/MS.
- Wide Dynamic Range: Up to 5 orders of magnitude dynamic range, suitable for both trace-level and bulk GAG quantification.
- Structural Elucidation: Isomer-specific identification of sulfation patterns and chain lengths with >95% structural coverage.
- High Throughput: Capable of processing up to 96 samples per batch with consistent performance metrics (CV<10%).
- Quantitative Accuracy: Employing internal standards and isotope-labeled analogs for absolute quantification (R² > 0.995).
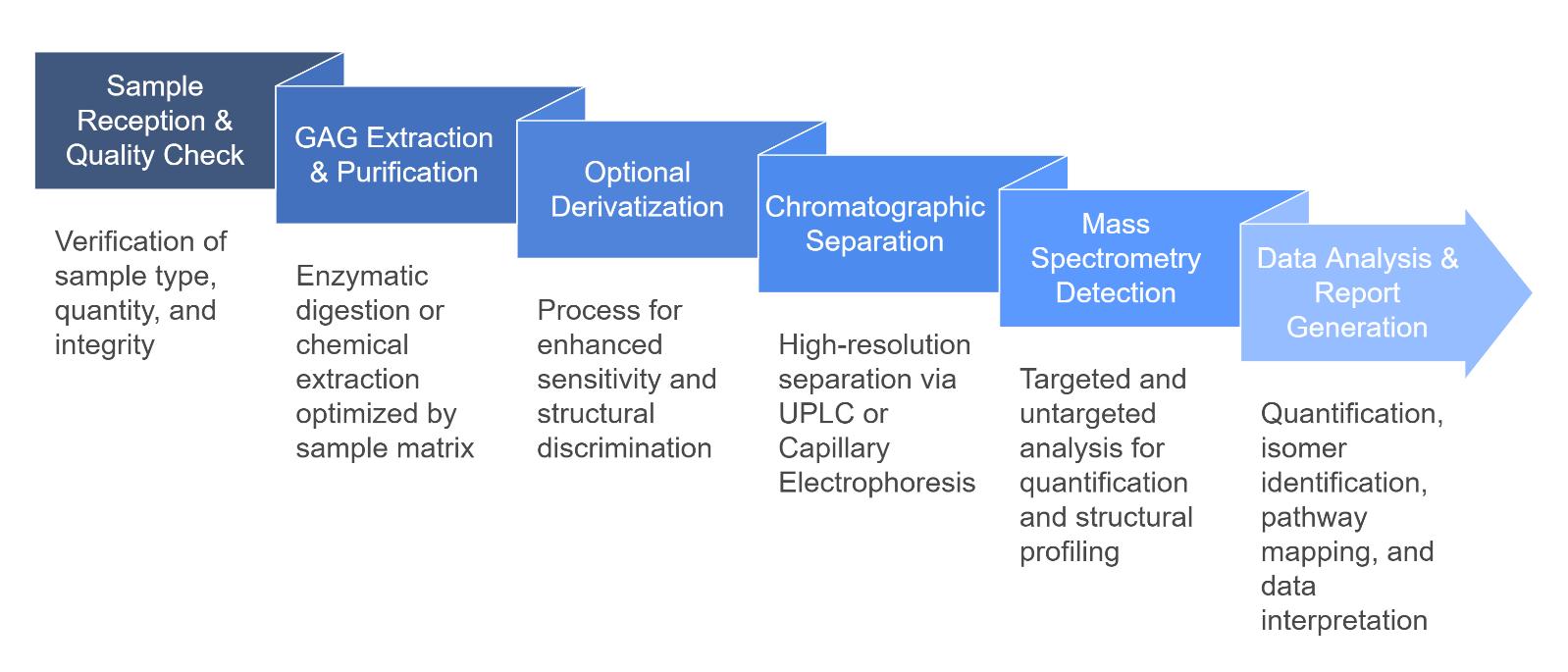
Workflow for Glycosaminoglycans Analysis Service
Technology Platform for Glycosaminoglycans Analysis Service
- UPLC-MS/MS (Waters ACQUITY UPLC + Xevo TQ-XS): For high-sensitivity quantitative analysis of GAG disaccharides and metabolites.
- High-Resolution LC-MS (Thermo Q Exactive Plus Orbitrap): For structural elucidation and detailed sulfation pattern profiling.

Waters ACQUITY UPLC System (Figure from Waters)

Thermo Fisher Q Exactive (Figure from Thermo Fisher)
Sample Requirements for Glycosaminoglycans Analysis Service
| Sample Type | Recommended Amount | Notes |
|---|---|---|
| Serum / Plasma | ≥ 100 µL | Collected with anticoagulants (e.g., EDTA); avoid hemolysis |
| Urine | ≥ 500 µL | First morning urine preferred; centrifuge before shipping |
| Tissue (animal/human) | ≥ 100 mg | Flash-frozen in liquid nitrogen; avoid repeated freeze-thaw cycles |
| Cell Pellets | ≥ 1 × 10⁷ cells | Washed with PBS; store at –80°C |
| CSF | ≥ 100 µL | Handle with care to prevent contamination |
| Cultured Medium | ≥ 1 mL | Serum-free preferred; filter before shipment |
| Feces | ≥ 200 mg | Collected in sterile containers; freeze-dried samples acceptable |
| Microbial Biomass | ≥ 500 mg (wet weight) or 10⁸ cells | Washed and flash-frozen; dry weight acceptable if freeze-dried |
| Other Biological Fluids | Contact us for evaluation | Custom sample types supported upon consultation |
Applications of Glycosaminoglycans Assay Service
Basic Biological Research
Investigating the role of GAGs in cellular processes, ECM dynamics, and cell-matrix interactions.
Drug Development Research
Analyzing GAG interactions with small molecules, proteins, and enzymes in therapeutic research.
Disease Mechanism Research
Studying GAG alterations in various disease models, such as cancer, fibrosis, and neurodegenerative diseases.
Tissue Engineering
Characterizing GAGs in scaffold development and cellular support for engineered tissues and implants.
Biomarker Discovery for Drug Development
Identifying GAG-related biomarkers to support early-stage drug discovery and development.
Biopharmaceutical Production
Ensuring the quality and consistency of GAG-based biologics, therapeutics, and vaccine development.
Demo
FAQ of Glycosaminoglycans Analysis Service
What is the sensitivity of the UPLC-MS/MS system for detecting GAG disaccharides?
The Waters ACQUITY UPLC + Xevo TQ-XS platform achieves detection limits as low as 0.1 pmol for targeted GAG-derived disaccharides, with a linear dynamic range spanning five orders of magnitude (e.g., 0.1–10,000 pmol). This ensures reliable quantification even in low-abundance biological matrices like plasma or cerebrospinal fluid.
How does the High-Resolution LC-MS system resolve complex sulfation patterns?
The Thermo Q Exactive Plus Orbitrap provides mass accuracy<2 ppm and resolving power up to 160,000 FWHM, enabling precise differentiation of sulfation isomers (e.g., ΔUA-GlcNS(6S) vs. ΔUA,2S-GlcNS). Its tandem MS capabilities generate high-quality MS/MS spectra for structural elucidation of hybrid GAG chains.
Can the workflow analyze both free GAGs and their metabolites?
Yes. The UPLC-MS/MS system quantifies free disaccharides (e.g., HA-derived ΔUA-GlcNAc), while the Orbitrap identifies sulfated metabolites (e.g., heparan sulfate oligosaccharides) through untargeted profiling. Combined, they cover >90% of known GAG-related metabolites in a single workflow.
What measures are taken to minimize matrix interference in complex samples?
- Ion-pairing chromatography (Waters ACQUITY UPLC) reduces co-elution of non-GAG contaminants.
- StepWave ion transfer optics (Xevo TQ-XS) actively eliminate neutral contaminants, enhancing signal-to-noise ratios by >50% in tissues or blood spots.
Can the Orbitrap system detect novel sulfated metabolites?
Yes. Its RADAR™ acquisition mode simultaneously collects quantitative MRM data and untargeted full-scan spectra, enabling discovery of uncharacterized sulfated metabolites (e.g., hybrid CS/KS structures) with >80% confidence.
What quality control (QC) standards are included?
- Internal standards: Isotopically labeled dermatan sulfate (DS-13C613C6) and keratan sulfate (KS-15N15N).
- System suitability tests: Daily injections of NIST SRM 3672 (chondroitin sulfate) to ensure CV<8% for retention time and intensity.
How are data reports structured for interdisciplinary teams?
Deliverables include:
- Excel files: Raw MRM transitions, peak areas, and CVs.
- PDF summaries: Heatmaps of sulfation patterns, pathway networks, and annotated MS/MS spectra.
Learn about other Q&A.
Glycosaminoglycans Analysis Service Case Study
Publications
Here are some publications in Metabolomics research from our clients:

- Identification of the O-Glycan Epitope Targeted by the Anti-Human Carcinoma Monoclonal Antibody (mAb) NEO-201. 2022. https://doi.org/10.3390/cancers14204999
- Modulation of the Endomembrane System by the Anticancer Natural Product Superstolide/ZJ-101. 2023. https://doi.org/10.3390/ijms24119575
- Alternative glycosylation controls endoplasmic reticulum dynamics and tubular extension in mammalian cells. 2021. https://doi.org/10.1126/sciadv.abe8349
- Conformational differences between functional human immunodeficiency virus envelope glycoprotein trimers and stabilized soluble trimers. 2019. https://doi.org/10.1128/jvi.01709-18
Reference
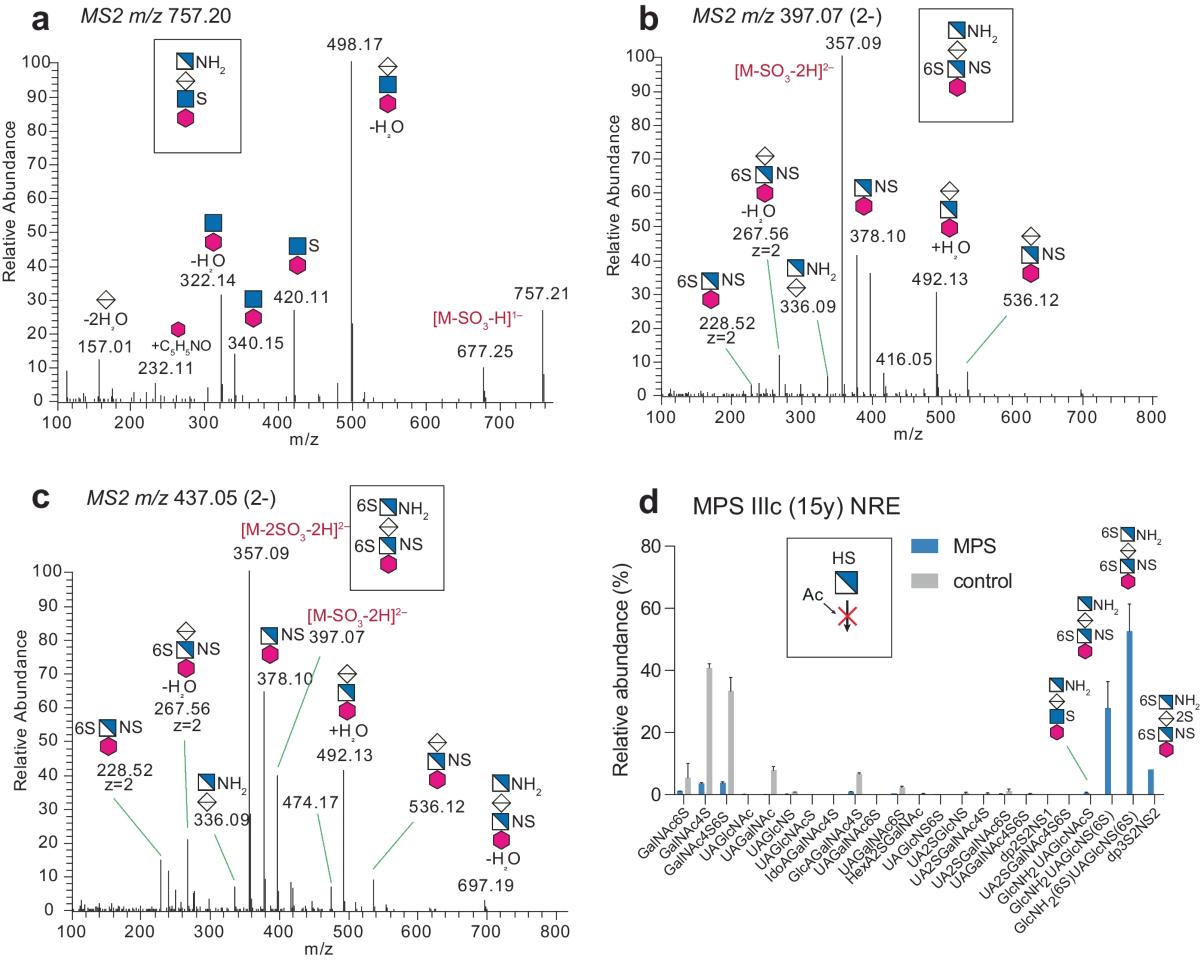
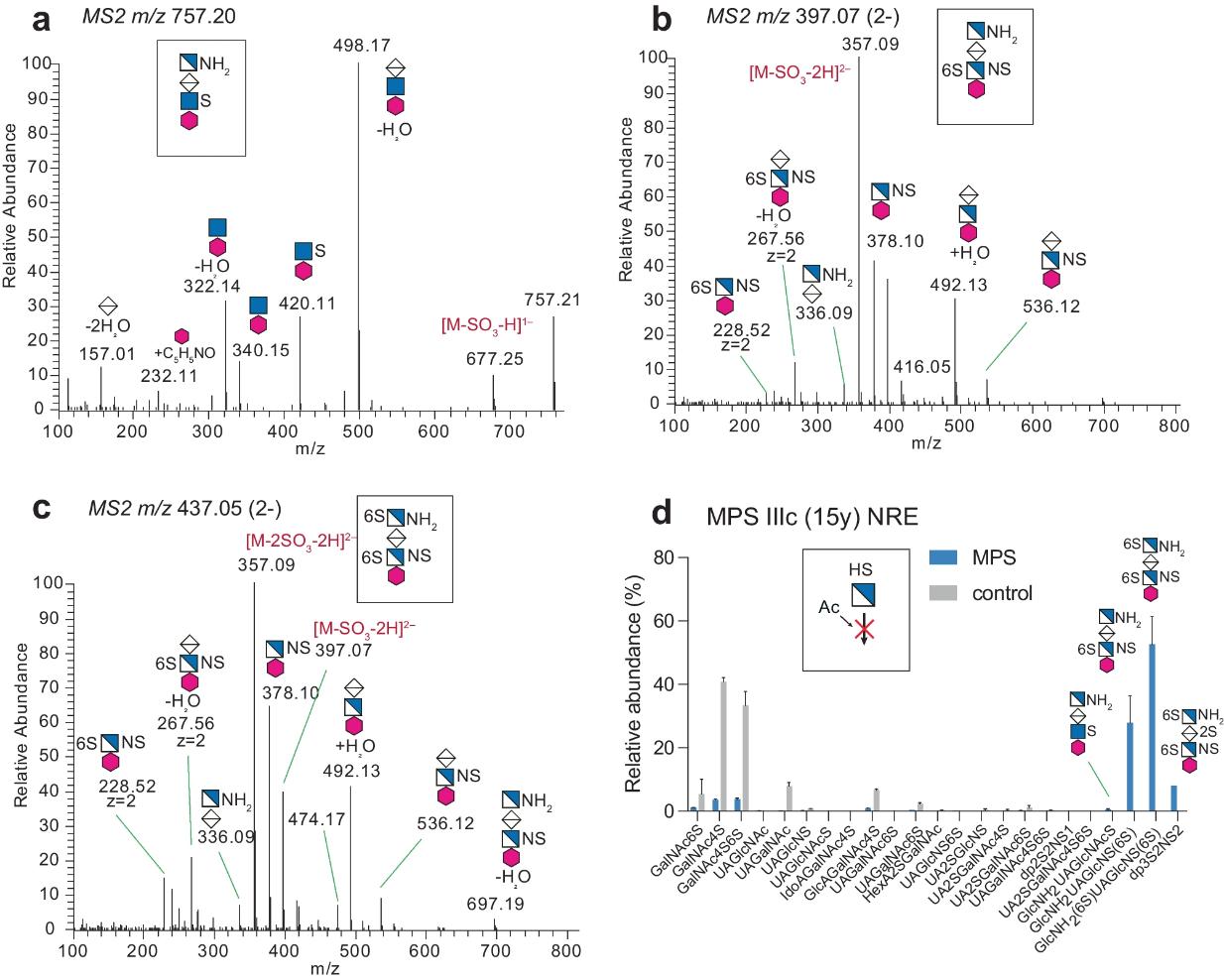
- Nilsson, Jonas, et al. "A glycomic workflow for LC–MS/MS analysis of urine glycosaminoglycan biomarkers in mucopolysaccharidoses." Glycoconjugate Journal 40.5 (2023): 523-540. https://doi.org/10.1007/s10719-023-10128-5