Diaminopimelic Acid (DAP) Analysis Service
At Creative Proteomics, we empower your research and industrial projects with highly sensitive, precise analysis of diaminopimelic acid (DAP). Whether you're tracking bacterial taxonomy, evaluating fermentation efficiency, or ensuring product quality, our specialized services deliver the data you need to move forward with confidence.
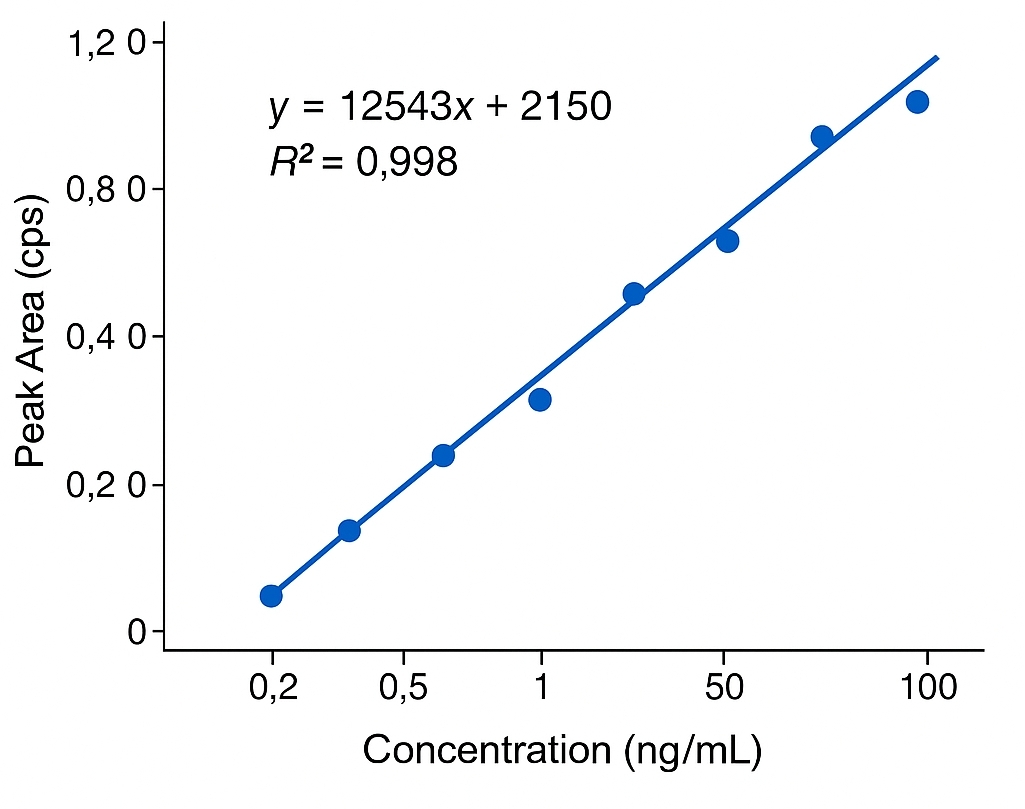
- Ultra-Low Detection Limits: Trace-level sensitivity down to ~0.2 ng/mL
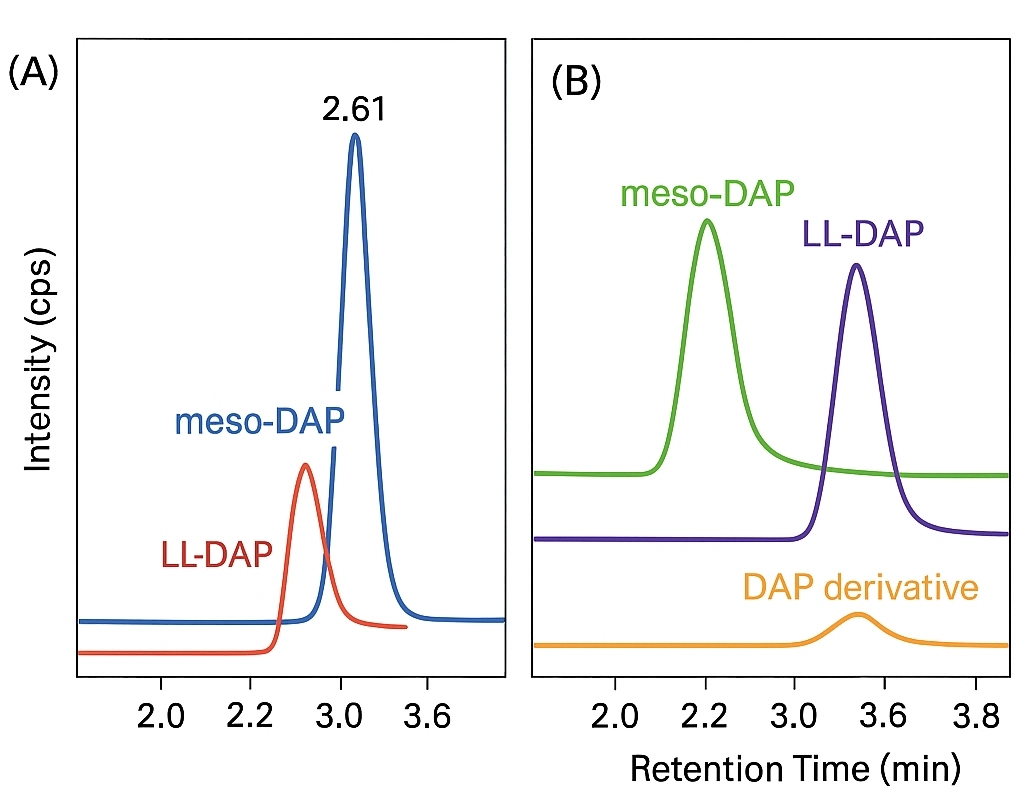
- Isomer-Specific Analysis: Distinguish meso-DAP and LL-DAP precisely
- Validated Across Matrices: Compatible with cultures, tissues, soil, and food
- Robust QC Standards: RSD<5%, strong="" linearity="">0.995)
- Expert Data Interpretation: Tailored insights for your specific goals
Submit Your Request Now
×
Deliverables You Can Expect:
- Quantitative DAP reports with isomer resolution
- High-quality chromatograms and spectra
- Validated method details for transparency
- Expert guidance to connect data with biological insights
- What We Provide
- Technology Platform
- Sample Requirement
- Demo
- Case
- FAQ
Why Diaminopimelic Acid Matters?
Diaminopimelic acid (DAP) is a pivotal component in bacterial cell wall biosynthesis and a critical chemotaxonomic marker for distinguishing Gram-positive and Gram-negative bacterial species. Beyond structural biology, its presence and quantitative levels have profound implications in microbiology, biotechnology, agricultural research, and the development of antimicrobial strategies.
Researchers and industrial partners alike demand high-sensitivity, high-accuracy analysis of DAP to:
- Track peptidoglycan synthesis and remodeling
- Profile microbial community composition in complex matrices
- Evaluate fermentation processes and bioengineering outcomes
- Investigate metabolic pathways relevant to microbial physiology and resistance mechanisms
Creative Proteomics offers a specialized diaminopimelic acid analysis service to deliver the depth of data required for high-level research and industrial applications.
Your Research Needs Drive Our Service Design
Need Ultra-Low Detection Limits for Trace Analysis?
→ We Deliver High Sensitivity and Precision
- LOD and LOQ: Down to low nanogram per milliliter levels, enabling trace-level detection in diverse matrices
- Instrument Platform: Advanced LC-MS/MS and GC-MS systems ensure robust sensitivity and signal fidelity
- Quantitation Range: Broad linear dynamic range for simultaneous low- and high-abundance measurements
- Reproducibility: RSD typically<5% across replicates
Our methods are optimized for complex samples—including bacterial cultures, fermentation broths, plant extracts, soil, and food matrices—to ensure confident detection even in highly variable backgrounds.
Need To Differentiate DAP Isomers or Derivatives?
→ We Provide Isomer-Specific Resolution
- Isomer Separation: Accurate differentiation between meso-DAP and LL-DAP, which is essential for distinguishing bacterial species or metabolic states
- Analytical Methods: Tailored derivatization techniques combined with chromatographic resolution for unambiguous identification
- Custom Method Development: For unique analytes or uncommon matrices, our scientists can develop bespoke workflows
Need Reliable Data for Downstream Analyses?
→ We Guarantee Data Quality and Compliance
- Method Validation: All methods are fully validated according to recognized analytical standards for:
- Linearity
- Precision
- Accuracy
- Recovery
- Matrix effects
- Quality Control: Includes calibration curves, blanks, spiked recoveries, and internal standards to ensure analytical integrity
- Documentation: Comprehensive reporting with method parameters, QC results, and quantitative data tables
Need Flexibility Across Diverse Applications?
→ We Support Multiple Research and Industrial Goals
Our DAP analysis services are widely applicable for:
- Microbial Taxonomy & Ecology: Profiling bacterial communities via chemotaxonomic markers
- Fermentation & Bioprocessing: Monitoring DAP as a marker of microbial metabolism in industrial fermentations
- Food Safety & Quality: Detecting bacterial contamination or fermentation by-products in food matrices
- Agricultural Studies: Investigating plant-microbe interactions and soil microbiomes
- Biotechnology R&D: Characterizing bacterial strains engineered for industrial applications
List of Detected DAP and Related Metabolites
| Compound | Biological Role | Primary Metabolic Pathways |
|---|---|---|
| meso-DAP (Diaminopimelic Acid) | Bacterial cell wall component; chemotaxonomic marker | Peptidoglycan synthesis, bacterial taxonomy |
| LL-DAP (Diaminopimelic Acid) | Isomeric form; species-specific biomarker | Bacterial cell wall synthesis, strain differentiation |
| DAP Derivatives | Modified forms used in metabolic studies | Peptidoglycan metabolism, bacterial physiology |
| NAD⁺ (Nicotinamide Adenine Dinucleotide) | Oxidized electron acceptor in redox reactions | Glycolysis, TCA Cycle, Oxidative Phosphorylation |
| NADH | Reduced electron donor, drives ATP synthesis | Electron Transport Chain, Respiration |
| NADP⁺ (Nicotinamide Adenine Dinucleotide Phosphate) | Oxidized coenzyme for biosynthesis | Pentose Phosphate Pathway, Fatty Acid Synthesis |
| NADPH | Reduced coenzyme for biosynthesis and antioxidant defense | Fatty Acid Synthesis, Glutathione Reduction |
| Niacin (Vitamin B3) | Dietary precursor for NAD⁺ biosynthesis | De Novo NAD⁺ Biosynthesis |
| Nicotinamide | Salvage pathway intermediate for NAD⁺ synthesis | NAD⁺ Salvage Pathway |
| Nicotinamide Riboside | Alternative NAD⁺ precursor | Nicotinamide Riboside Kinase Pathway |
| Nicotinamide Mononucleotide (NMN) | Direct NAD⁺ precursor | NAD⁺ Biosynthesis Intermediate |
| Nicotinic Acid Adenine Dinucleotide (NAAD) | Intermediate in NAD⁺ biosynthesis | Preiss-Handler NAD⁺ Salvage Pathway |
| Coenzyme Q10 (Ubiquinone) | Lipid-soluble electron carrier in mitochondria | Mitochondrial Respiratory Chain (Complex I–III) |
Methods and Instrumentation for DAP Analysis
LC-MS/MS Analysis
- System: Agilent 6495C Triple Quadrupole LC/MS
- Column: Waters XBridge BEH C18, 2.1 × 100 mm, 1.7 μm
- Ionization: ESI Positive Mode
- LOD: ~0.2 ng/mL
- Strengths: Exceptional sensitivity, isomer resolution, suited for complex matrices
 Agilent 6495C Triple quadrupole (Figure from Agilent)
Agilent 6495C Triple quadrupole (Figure from Agilent)
GC-MS Analysis
- System: Agilent 7890B GC coupled with 5977A MSD
- Derivatization: BSTFA
- Column: DB-5MS UI, 30 m × 0.25 mm × 0.25 μm
- Strengths: Ideal for volatile derivatives and robust QC applications
 Agilent 7890B-5977A (Figure from Agilent)
Agilent 7890B-5977A (Figure from Agilent)
HPLC-FLD Analysis
- System: Agilent 1260 Infinity II HPLC with fluorescence detector
- Derivatization: o-Phthalaldehyde (OPA) or FMOC-Cl
- Excitation/Emission: 340/455 nm (OPA derivatives)
- Strengths: Cost-effective, reliable for routine analysis
 Agilent 1260 Infinity II HPLC (Figure from Agilent)
Agilent 1260 Infinity II HPLC (Figure from Agilent)
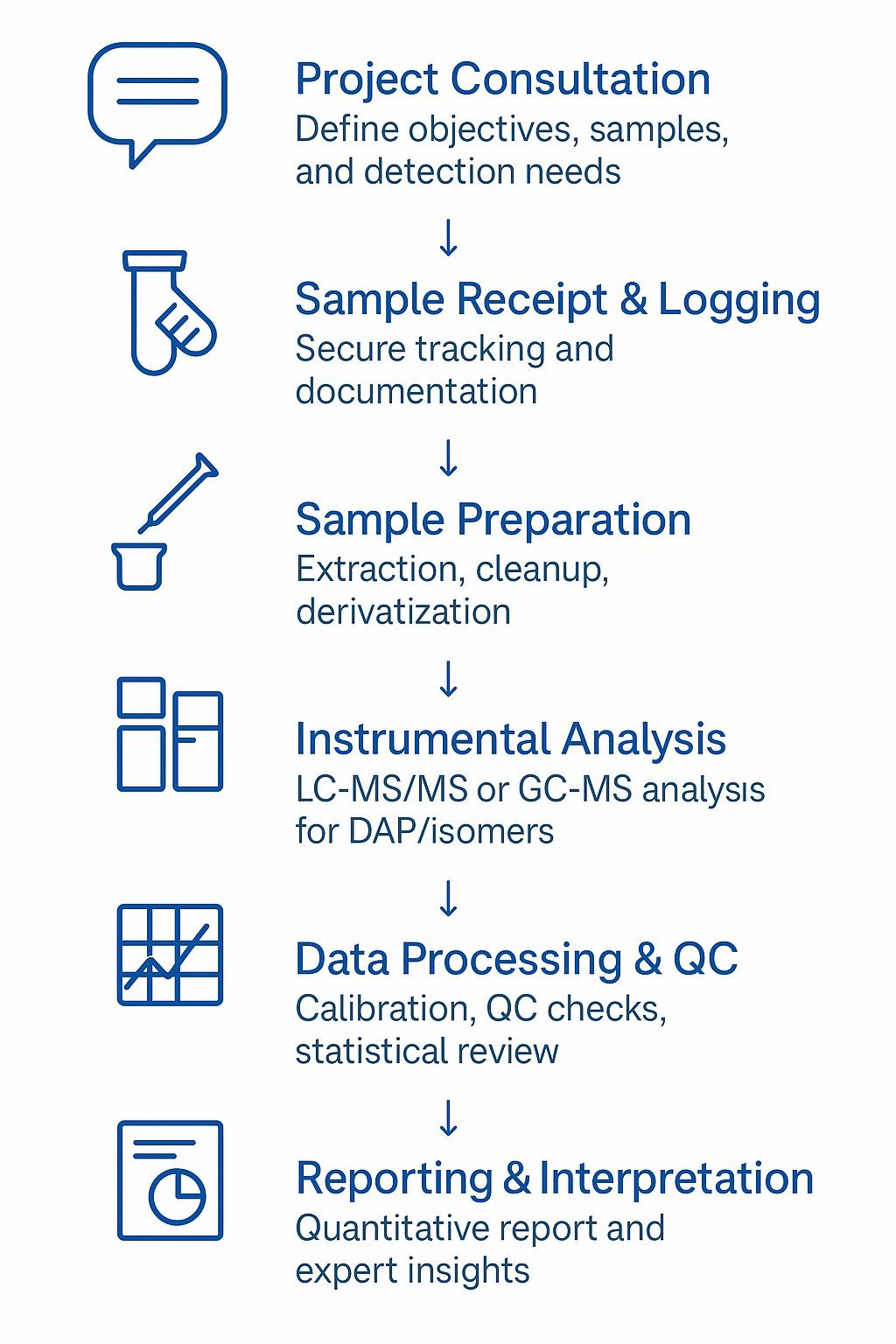
Analytical Workflow: Your Path to Reliable DAP Data
1. Project Consultation
- Define research objectives, sample types, and detection requirements
2. Sample Receipt & Logging
- Secure tracking and documentation of all incoming samples
3. Sample Preparation
- Matrix-specific extraction and clean-up
- Derivatization (e.g., o-phthalaldehyde, FMOC) for enhanced detection sensitivity
4. Instrumental Analysis
- LC-MS/MS or GC-MS platform selection based on matrix and detection goals
- Optimized gradient and MRM transitions for DAP and its isomers
5. Data Processing & QC
- Calibration curve fitting
- Validation of internal standard performance
- Statistical verification of replicates
6. Reporting & Interpretation
- Comprehensive report with absolute quantification data
- Optional interpretation assistance to connect results to biological pathways or industrial process control
Sample Requirements for DAP Analysis Service
| Sample Type | Required Amount / Volume | Storage Condition | Notes |
|---|---|---|---|
| Bacterial Cultures | ≥ 5 mL | -80°C | Pellet cells if possible before freezing |
| Fermentation Broth | ≥ 2 mL | -20°C or -80°C | Remove particulates if feasible |
| Plant Tissue | ≥ 50 mg | -80°C | Snap-freeze after collection |
| Soil Samples | ≥ 1 g | -20°C or -80°C | Air-dry or freeze-dry recommended |
| Animal Tissue | ≥ 50 mg | -80°C | Snap-freeze after dissection |
| Food Samples | ≥ 1 g | -20°C or -80°C | Homogenize thoroughly |
| Cell Pellets | ≥ 1 × 10⁶ cells | -80°C | Wash with PBS before freezing |
Demo Results
DAP Analysis Service Case Study

Title: Integrated Transcriptome and Metabolome Analysis Reveals Key Metabolic Pathways in Mulberry Response to Botrytis cinerea Infection
Journal: BMC Plant Biology
Published:2024
- Study Summary
- Background
- Methods
- Results
- Reference
This study investigated the molecular defense mechanisms of mulberry (Morus alba L.) in response to infection by Botrytis cinerea. Through integrated transcriptome and metabolome profiling, the researchers identified critical genes and metabolic pathways activated during fungal invasion. A notable finding was the involvement of the diaminopimelic acid (DAP) pathway, which may play a role in plant immune responses.
Botrytis cinerea is a widespread necrotrophic pathogen responsible for gray mold in a variety of crops, including mulberry. The infection significantly impacts leaf health and crop yield. To better understand mulberry's defense responses, the authors performed a time-course omics study combining RNA-seq and LC-MS-based metabolomics on infected leaves.
In this study, pathway enrichment analysis based on transcriptome and metabolome data revealed significant activation of the Diaminopimelic Acid (DAP) biosynthesis pathway, which is traditionally known for its role in bacterial cell wall and lysine biosynthesis. The researchers observed upregulation of genes such as MaLYS1 and MaALD1, indicating that the DAP pathway may be involved in the mulberry's defense mechanism against B. cinerea. These findings suggest an intriguing link between DAP metabolism and plant immunity.
![]() Creative Proteomics offers both targeted and untargeted DAP analysis services to support similar research needs. Our capabilities include:
Creative Proteomics offers both targeted and untargeted DAP analysis services to support similar research needs. Our capabilities include:
- Ultra-sensitive quantification of DAP-related metabolites such as NAD⁺, NADH, nicotinamide, nicotinate, NMN, and NR
- Advanced LC-MS/MS platforms (e.g., Agilent 6495C Triple Quadrupole) achieving LODs as low as 0.01 ng/mL
- Comprehensive pathway enrichment analysis linking DAP fluctuations to metabolic stress responses
- Customized method development for challenging sample types such as plant tissues
SAP induces gut barrier disruption and microbiota dysbiosis: SAP rats showed increased intestinal permeability and altered microbiota composition (↑ Firmicutes, Helicobacter, Lactobacillus; ↓ Bacteroidetes, Allobaculum).
Elevated serum DAP activates the NOD1/RIP2 pathway: DAP levels were significantly higher in SAP rats as detected by UHPLC-TQMS, correlating with increased expression of NOD1, RIP2, and phosphorylated NF-κB, indicating innate immune activation.
DAP serves as an early biomarker in clinical SAP: In patients, serum DAP levels were significantly elevated in severe AP cases, with strong diagnostic potential (AUC = 0.82), independent of other clinical features.
Interventions reduce DAP and suppress inflammation: Both QYKL and Neomycin treatment lowered serum DAP levels, restored gut microbiota balance, and inhibited the NOD1/RIP2 inflammatory signaling cascade, mitigating systemic inflammation.
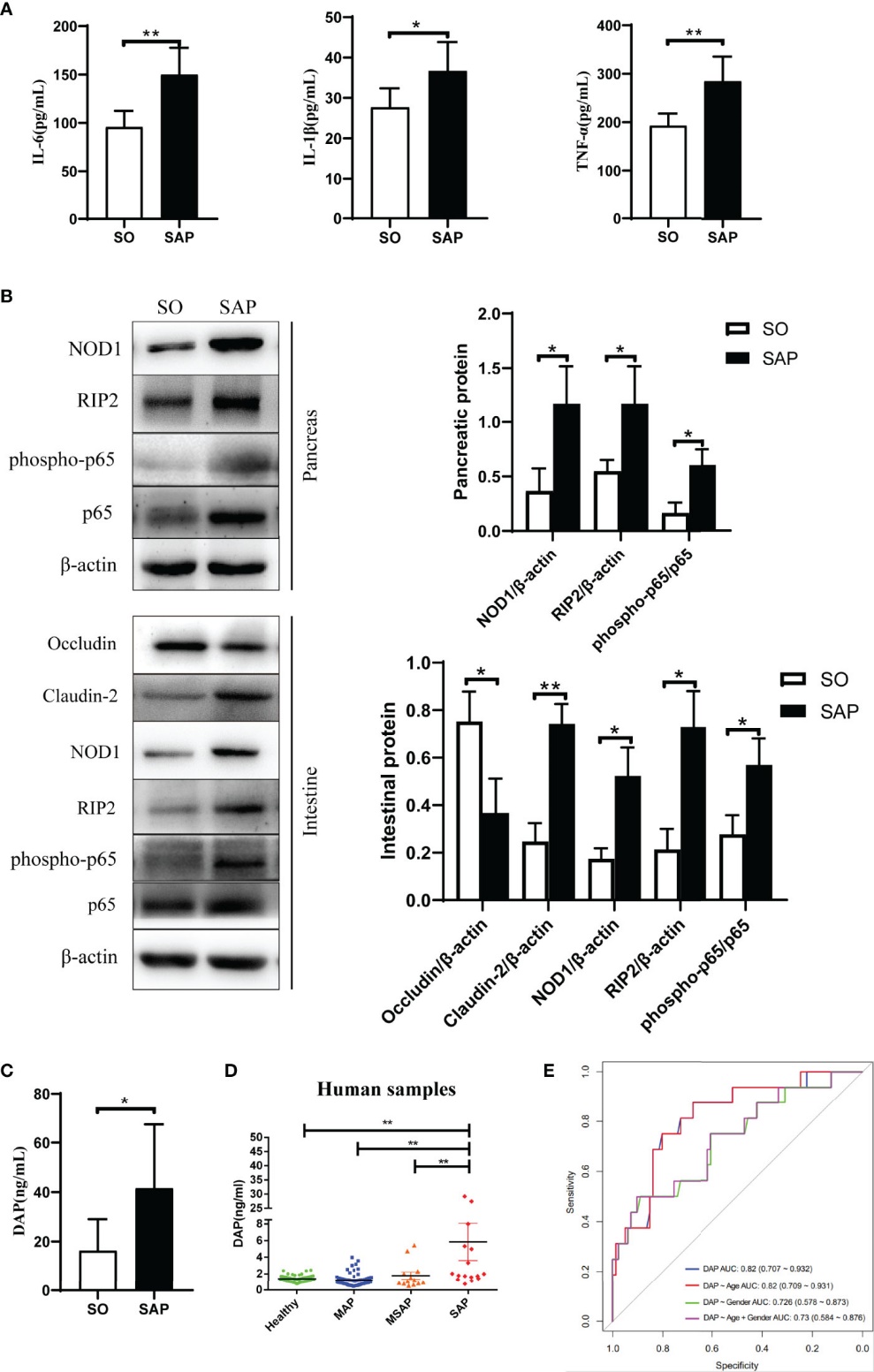 DAP activates the NOD1/RIP2 pathway and drives inflammation in SAP rats, with elevated serum DAP in SAP patients. (A) Serum IL-6, IL-1β, TNF-α in SO vs. SAP rats. (B) NOD1 pathway and barrier protein levels in pancreas and intestine. (C) Serum DAP in SO and SAP rats. (D) Serum DAP by AP severity in patients. (E) ROC for serum DAP distinguishing SAP from MAP/MSAP. *p < 0.05, **p < 0.01.
DAP activates the NOD1/RIP2 pathway and drives inflammation in SAP rats, with elevated serum DAP in SAP patients. (A) Serum IL-6, IL-1β, TNF-α in SO vs. SAP rats. (B) NOD1 pathway and barrier protein levels in pancreas and intestine. (C) Serum DAP in SO and SAP rats. (D) Serum DAP by AP severity in patients. (E) ROC for serum DAP distinguishing SAP from MAP/MSAP. *p < 0.05, **p < 0.01.
Reference
- Jiao, Juying, et al. "Gut microbiota-derived diaminopimelic acid promotes the NOD1/RIP2 signaling pathway and plays a key role in the progression of severe acute pancreatitis." Frontiers in Cellular and Infection Microbiology 12 (2022): 838340. https://doi.org/10.3389/fcimb.2022.838340
FAQ of DAP Analysis Service
Can you distinguish between meso-DAP and LL-DAP in your analysis?
Yes. Our LC-MS/MS method is optimized to resolve meso- and LL- isomers of DAP by using specific chromatographic separation and targeted MRM transitions, ensuring precise identification and quantification.
What matrices have you validated for DAP detection?
Our assays are validated for a wide range of matrices including bacterial cultures, fermentation broths, plant tissues, soil extracts, food samples, and cell pellets. For unusual matrices, we offer method adaptation and validation upon request.
How do you handle matrix effects in complex samples?
We employ matrix-matched calibration curves, stable isotope-labeled internal standards (if available), and rigorous sample cleanup procedures to minimize ion suppression or enhancement, ensuring accurate quantification in complex sample types.
Can you accommodate low-concentration samples with minimal biomass or volume?
Absolutely. Our instrumentation achieves detection limits as low as ~0.2 ng/mL. We regularly analyze low-volume or low-biomass samples by adjusting extraction and concentration workflows to maximize recovery.
Is it possible to integrate DAP analysis results with other metabolomics data?
Definitely. We can deliver your data in formats compatible with multi-omics platforms and support integration with metabolomics, proteomics, or microbiome datasets for systems-level analysis.
Can DAP quantification support microbial taxonomy studies?
Yes. DAP composition and stereochemistry are critical chemotaxonomic markers for differentiating bacterial species and can complement genomic or proteomic data in taxonomic studies.
Do you offer custom method development for unique research applications?
Yes. We frequently develop customized analytical methods for clients with specialized research goals, unique matrices, or regulatory needs. Contact us to discuss feasibility and requirements.
Learn about other Q&A about proteomics technology.
Publications
Here are some of the metabolomics-related papers published by our clients:

- Methyl donor supplementation reduces phospho‐Tau, Fyn and demethylated protein phosphatase 2A levels and mitigates learning and motor deficits in a mouse model of tauopathy. 2023. https://doi.org/10.1111/nan.12931
- A human iPSC-derived hepatocyte screen identifies compounds that inhibit production of Apolipoprotein B. 2023. https://doi.org/10.1038/s42003-023-04739-9
- The activity of the aryl hydrocarbon receptor in T cells tunes the gut microenvironment to sustain autoimmunity and neuroinflammation. 2023. https://doi.org/10.1371/journal.pbio.3002000
- Lipid droplet-associated lncRNA LIPTER preserves cardiac lipid metabolism. 2023. https://doi.org/10.1038/s41556-023-01162-4
- Inflammation primes the kidney for recovery by activating AZIN1 A-to-I editing. 2023. https://doi.org/10.1101/2023.11.09.566426
- Anti-inflammatory activity of black soldier fly oil associated with modulation of TLR signaling: A metabolomic approach. 2023. https://doi.org/10.3390/ijms241310634
- Plant Growth Promotion, Phytohormone Production and Genomics of the Rhizosphere-Associated Microalga, Micractinium rhizosphaerae sp. 2023. https://doi.org/10.3390/plants12030651